Favipiravir-resistant influenza A virus shows potential for transmission
- PMID: 34061908
- PMCID: PMC8195362
- DOI: 10.1371/journal.ppat.1008937
Favipiravir-resistant influenza A virus shows potential for transmission
Abstract
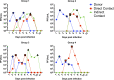
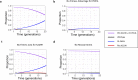
Favipiravir is a nucleoside analogue which has been licensed to treat influenza in the event of a new pandemic. We previously described a favipiravir resistant influenza A virus generated by in vitro passage in presence of drug with two mutations: K229R in PB1, which conferred resistance at a cost to polymerase activity, and P653L in PA, which compensated for the cost of polymerase activity. However, the clinical relevance of these mutations is unclear as the mutations have not been found in natural isolates and it is unknown whether viruses harbouring these mutations would replicate or transmit in vivo. Here, we infected ferrets with a mix of wild type p(H1N1) 2009 and corresponding favipiravir-resistant virus and tested for replication and transmission in the absence of drug. Favipiravir-resistant virus successfully infected ferrets and was transmitted by both contact transmission and respiratory droplet routes. However, sequencing revealed the mutation that conferred resistance, K229R, decreased in frequency over time within ferrets. Modelling revealed that due to a fitness advantage for the PA P653L mutant, reassortment with the wild-type virus to gain wild-type PB1 segment in vivo resulted in the loss of the PB1 resistance mutation K229R. We demonstrated that this fitness advantage of PA P653L in the background of our starting virus A/England/195/2009 was due to a maladapted PA in first wave isolates from the 2009 pandemic. We show there is no fitness advantage of P653L in more recent pH1N1 influenza A viruses. Therefore, whilst favipiravir-resistant virus can transmit in vivo, the likelihood that the resistance mutation is retained in the absence of drug pressure may vary depending on the genetic background of the starting viral strain.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: WB has received honoraria from Roche, Sanofi Pasteur and Seqirus. The rest of the Authors have nothing to declare.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

