Anticipating response function in gene regulatory networks
- PMID: 34062105
- PMCID: PMC8169214
- DOI: 10.1098/rsif.2021.0206
Anticipating response function in gene regulatory networks
Abstract
The origin of an ordered genetic response of a complex and noisy biological cell is intimately related to the detailed mechanism of protein-DNA interactions present in a wide variety of gene regulatory (GR) systems. However, the quantitative prediction of genetic response and the correlation between the mechanism and the response curve is poorly understood. Here, we report in silico binding studies of GR systems to show that the transcription factor (TF) binds to multiple DNA sites with high cooperativity spreads from specific binding sites into adjacent non-specific DNA and bends the DNA. Our analysis is not limited only to the isolated model system but also can be applied to a system containing multiple interacting genes. The controlling role of TF oligomerization, TF-ligand interactions, and DNA looping for gene expression has been also characterized. The predictions are validated against detailed grand canonical Monte Carlo simulations and published data for the lac operon system. Overall, our study reveals that the expression of target genes can be quantitatively controlled by modulating TF-ligand interactions and the bending energy of DNA.
Keywords: GCMC simulation; gene regulation; protein–DNA networks; response function; statistical mechanics.
Figures

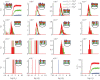
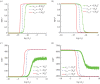
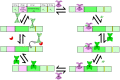
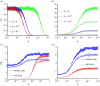
Similar articles
-
Self-consistent theory of transcriptional control in complex regulatory architectures.PLoS One. 2017 Jul 7;12(7):e0179235. doi: 10.1371/journal.pone.0179235. eCollection 2017. PLoS One. 2017. PMID: 28686609 Free PMC article.
-
A biophysical model for analysis of transcription factor interaction and binding site arrangement from genome-wide binding data.PLoS One. 2009 Dec 1;4(12):e8155. doi: 10.1371/journal.pone.0008155. PLoS One. 2009. PMID: 19956545 Free PMC article.
-
Determinants of correlated expression of transcription factors and their target genes.Nucleic Acids Res. 2020 Nov 18;48(20):11347-11369. doi: 10.1093/nar/gkaa927. Nucleic Acids Res. 2020. PMID: 33104784 Free PMC article.
-
Ordering up gene expression by slowing down transcription factor binding kinetics.Curr Genet. 2019 Apr;65(2):401-406. doi: 10.1007/s00294-018-0896-7. Epub 2018 Oct 23. Curr Genet. 2019. PMID: 30353359 Free PMC article. Review.
-
Identification of plant transcription factor target sequences.Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):21-30. doi: 10.1016/j.bbagrm.2016.05.001. Epub 2016 May 4. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27155066 Review.
Cited by
-
In vivo, in vitro and in silico: an open space for the development of microbe-based applications of synthetic biology.Microb Biotechnol. 2022 Jan;15(1):42-64. doi: 10.1111/1751-7915.13937. Epub 2021 Sep 27. Microb Biotechnol. 2022. PMID: 34570957 Free PMC article.
-
The Blueprint of Logical Decisions in a NF-κB Signaling System.ACS Omega. 2024 May 13;9(21):22625-22634. doi: 10.1021/acsomega.4c00049. eCollection 2024 May 28. ACS Omega. 2024. PMID: 38826544 Free PMC article.
References
-
- Alberts B. 2017. Molecular biology of the cell. New York, NY: CRC Press.
-
- Phillips R, Theriot J, Kondev J, Garcia H. 2012. Physical biology of the cell. New York, NY:Garland Science.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

