Chemical system biology approach to identify multi-targeting FDA inhibitors for treating COVID-19 and associated health complications
- PMID: 34062110
- PMCID: PMC8171008
- DOI: 10.1080/07391102.2021.1931451
Chemical system biology approach to identify multi-targeting FDA inhibitors for treating COVID-19 and associated health complications
Abstract
In view of many European countries and the USA leading to the second wave of COVID-19 pandemic, winter season, the evolution of new mutations in the spike protein, and no registered drugs and vaccines for COVID-19 treatment, the discovery of effective and novel therapeutic agents is urgently required. The degrees and frequencies of COVID-19 clinical complications are related to uncontrolled immune responses, secondary bacterial infections, diabetes, cardiovascular disease, hypertension, and chronic pulmonary diseases. It is essential to recognize that the drug repurposing strategy so far remains the only means to manage the disease burden of COVID-19. Despite some success of using single-target drugs in treating the disease, it is beyond suspicion that the virus will acquire drug resistance by acquiring mutations in the drug target. The possible synergistic inhibition of drug efficacy due to drug-drug interaction cannot be avoided while treating COVID-19 and allied clinical complications. Hence, to avoid the unintended development drug resistance and loss of efficacy due to drug-drug interaction, multi-target drugs can be promising tools for the most challenging disease. In the present work, we have carried out molecular docking studies of compounds from the FDA approved drug library, and the FDA approved and passed phase -1 drug libraries with ten therapeutic targets of COVID-19. Results showed that known drugs, including nine anti-inflammatory compounds, four antibiotics, six antidiabetic compounds, and one cardioprotective compound, could effectively inhibit multiple therapeutic targets of COVID-19. Further in-vitro, in vivo, and clinical studies will guide these drugs' proper allocation to treat COVID-19.Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; FDA/passed phase-1 inhibitors; molecular docking; molecular dynamics; multi-targeting compounds.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
The authors have declared no competing interest.
Figures


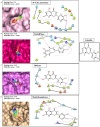


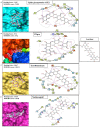
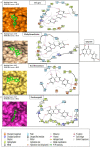
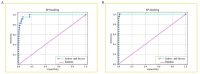
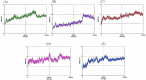
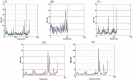
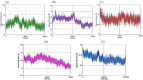
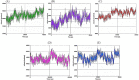
References
-
- Aishwarya, S., Gunasekaran, K., Sagaya Jansi, R., & Sangeetha, G. (2021). From genomes to molecular dynamics- A bottom up approach in extrication of SARS CoV-2 main protease inhibitors. Computational Toxicology (Amsterdam, Netherlands), 18, 100156. https://doi.org/ 10.1016/j.comtox.2021.100156 - DOI - PMC - PubMed
-
- Akriti, K., Satpathy, I., & Patnaik, B. (2021). Covid-19 and its impact on livelihood: An Indian perspective. Eurasian Chemical Communications, 3(2), 81–87.
-
- Altayeb, H., Bouslama, L., Abdulhakimc, J. A., Chaieb, K., Baothman, O. A., & Zamzami, M. A. (2020). Potential activity of a selected natural compounds on SARS-CoV-2 RNA-dependent-RNA polymerase, and binding affinity of the receptor-binding domain (RBD).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
