TRIM7 inhibits enterovirus replication and promotes emergence of a viral variant with increased pathogenicity
- PMID: 34062120
- PMCID: PMC8276836
- DOI: 10.1016/j.cell.2021.04.047
TRIM7 inhibits enterovirus replication and promotes emergence of a viral variant with increased pathogenicity
Abstract
To control viral infection, vertebrates rely on both inducible interferon responses and less well-characterized cell-intrinsic responses composed of "at the ready" antiviral effector proteins. Here, we show that E3 ubiquitin ligase TRIM7 is a cell-intrinsic antiviral effector that restricts multiple human enteroviruses by targeting viral 2BC, a membrane remodeling protein, for ubiquitination and proteasome-dependent degradation. Selective pressure exerted by TRIM7 results in emergence of a TRIM7-resistant coxsackievirus with a single point mutation in the viral 2C ATPase/helicase. In cultured cells, the mutation helps the virus evade TRIM7 but impairs optimal viral replication, and this correlates with a hyperactive and structurally plastic 2C ATPase. Unexpectedly, the TRIM7-resistant virus has a replication advantage in mice and causes lethal pancreatitis. These findings reveal a unique mechanism for targeting enterovirus replication and provide molecular insight into the benefits and trade-offs of viral evolution imposed by a host restriction factor.
Keywords: Antiviral immunity; E3 ubiquitin ligase; Enterovirus; Restriction factor; Viral evolution; Viral pathogenesis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





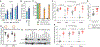
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, and Lindah E (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–15.
-
- Amadei A, Linssen ABM, and Berendsen HJC (1993). Essential dynamics of proteins. Proteins Struct. Funct. Bioinforma 17(4), 412–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

