SARS-CoV-2 RNA Quantification Using Droplet Digital RT-PCR
- PMID: 34062285
- PMCID: PMC8164350
- DOI: 10.1016/j.jmoldx.2021.04.014
SARS-CoV-2 RNA Quantification Using Droplet Digital RT-PCR
Abstract
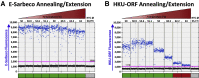
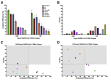
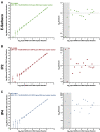
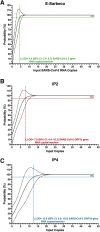
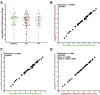
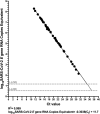
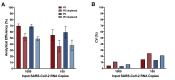
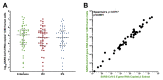
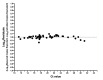
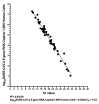
Quantitative viral load assays have transformed our understanding of viral diseases. They hold similar potential to advance COVID-19 control and prevention, but SARS-CoV-2 viral load tests are not yet widely available. SARS-CoV-2 molecular diagnostic tests, which typically employ real-time RT-PCR, yield semiquantitative results only. Droplet digital RT-PCR (RT-ddPCR) offers an attractive platform for SARS-CoV-2 RNA quantification. Eight primer/probe sets originally developed for real-time RT-PCR-based SARS-CoV-2 diagnostic tests were evaluated for use in RT-ddPCR; three were identified as the most efficient, precise, and sensitive for RT-ddPCR-based SARS-CoV-2 RNA quantification. For example, the analytical efficiency for the E-Sarbeco primer/probe set was approximately 83%, whereas assay precision, measured as the coefficient of variation, was approximately 2% at 1000 input copies/reaction. Lower limits of quantification and detection for this primer/probe set were 18.6 and 4.4 input SARS-CoV-2 RNA copies/reaction, respectively. SARS-CoV-2 RNA viral loads in a convenience panel of 48 COVID-19-positive diagnostic specimens spanned a 6.2log10 range, confirming substantial viral load variation in vivo. RT-ddPCR-derived SARS-CoV-2 E gene copy numbers were further calibrated against cycle threshold values from a commercial real-time RT-PCR diagnostic platform. This log-linear relationship can be used to mathematically derive SARS-CoV-2 RNA copy numbers from cycle threshold values, allowing the wealth of available diagnostic test data to be harnessed to address foundational questions in SARS-CoV-2 biology.
Copyright © 2021 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Mellors J.W., Rinaldo C.R., Jr., Gupta P., White R.M., Todd J.A., Kingsley L.A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. - PubMed
-
- Mellors J.W. Viral-load tests provide valuable answers. Sci Am. 1998;279:90–93. - PubMed
-
- Riddler S.A., Mellors J.W. HIV-1 viral dynamics and viral load measurement: implications for therapy. AIDS Clin Rev. 1997:47–65. - PubMed
-
- Mellors J.W., Muñoz A., Giorgi J.V., Margolick J.B., Tassoni C.J., Gupta P., Kingsley L.A., Todd J.A., Saah A.J., Detels R., Phair J.P., Rinaldo C.R., Jr. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. - PubMed
-
- Durante-Mangoni E., Zampino R., Portella G., Adinolfi L.E., Utili R., Ruggiero G. Correlates and prognostic value of the first-phase hepatitis C virus RNA kinetics during treatment. Clin Infect Dis. 2009;49:498–506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

