Electrophoretic mobility shift as a molecular beacon-based readout for miRNA detection
- PMID: 34062334
- PMCID: PMC8461749
- DOI: 10.1016/j.bios.2021.113307
Electrophoretic mobility shift as a molecular beacon-based readout for miRNA detection
Abstract
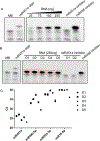
MicroRNAs are short, non-coding RNA sequences involved in gene expression regulation. Quantification of miRNAs in biological fluids involves time consuming and laborious methods such as Northern blotting or PCR-based techniques. Molecular beacons (MB) are an attractive means for rapid detection of miRNAs, although the need for sophisticated readout methods limits their use in research and clinical settings. Here, we introduce a novel method based on delayed electrophoretic mobility, as a quantitative means for detection of miRNAs-MB hybridization. Upon hybridization with the target miRNAs, MB form a fluorescent duplex with reduced electrophoretic mobility, thus bypassing the need for additional staining. In addition to emission of light, the location of the fluorescent band on the gel acts as an orthogonal validation of the target identity, further confirming the specificity of binding. The limit of detection of this approach is approximately 100 pM, depending on the MB sequence. The method is sensitive enough to detect specific red blood cell miRNAs molecules in total RNA, with single nucleotide specificity. Altogether, we describe a rapid and affordable method that offers sensitive detection of single-stranded small DNA and RNA sequences.
Keywords: Electrophoretic mobility; Gel electrophoresis; Molecular beacons; microRNA (miRNA).
Copyright © 2021 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
Beth Israel Deaconess Medical Center (GPO, ICG), and Boston Children’s Hospital (WPW) have submitted a patent covering the methods described in this manuscript. The other co-authors declare no conflict of interest.
Figures
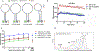
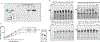
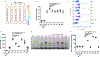
Similar articles
-
The use of molecular beacons to detect and quantify microRNA.Methods Mol Biol. 2013;1039:279-87. doi: 10.1007/978-1-62703-535-4_22. Methods Mol Biol. 2013. PMID: 24026703
-
A simple molecular beacon with duplex-specific nuclease amplification for detection of microRNA.Analyst. 2016 Feb 7;141(3):1071-6. doi: 10.1039/c5an02312b. Epub 2015 Dec 21. Analyst. 2016. PMID: 26688865
-
In vitro quantification of specific microRNA using molecular beacons.Nucleic Acids Res. 2012 Jan;40(2):e13. doi: 10.1093/nar/gkr1016. Epub 2011 Nov 21. Nucleic Acids Res. 2012. PMID: 22110035 Free PMC article.
-
A Cascade Signal Amplification Based on Dynamic DNA Nanodevices and CRISPR/Cas12a Trans-cleavage for Highly Sensitive MicroRNA Sensing.ACS Synth Biol. 2021 Jun 18;10(6):1481-1489. doi: 10.1021/acssynbio.1c00064. Epub 2021 May 19. ACS Synth Biol. 2021. PMID: 34011151
-
Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell.Acc Chem Res. 2017 Apr 18;50(4):1059-1068. doi: 10.1021/acs.accounts.7b00040. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28355077 Review.
Cited by
-
Nanopore-based detection of periodontitis biomarker miR31 in saliva samples.Electrophoresis. 2024 Nov;45(21-22):2034-2044. doi: 10.1002/elps.202400134. Epub 2024 Aug 21. Electrophoresis. 2024. PMID: 39165194
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

