Hippocampal Subregion and Gene Detection in Alzheimer's Disease Based on Genetic Clustering Random Forest
- PMID: 34062866
- PMCID: PMC8147351
- DOI: 10.3390/genes12050683
Hippocampal Subregion and Gene Detection in Alzheimer's Disease Based on Genetic Clustering Random Forest
Abstract
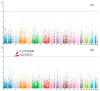
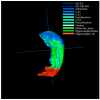
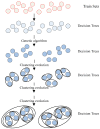
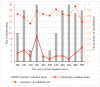
The distinguishable subregions that compose the hippocampus are differently involved in functions associated with Alzheimer's disease (AD). Thus, the identification of hippocampal subregions and genes that classify AD and healthy control (HC) groups with high accuracy is meaningful. In this study, by jointly analyzing the multimodal data, we propose a novel method to construct fusion features and a classification method based on the random forest for identifying the important features. Specifically, we construct the fusion features using the gene sequence and subregions correlation to reduce the diversity in same group. Moreover, samples and features are selected randomly to construct a random forest, and genetic algorithm and clustering evolutionary are used to amplify the difference in initial decision trees and evolve the trees. The features in resulting decision trees that reach the peak classification are the important "subregion gene pairs". The findings verify that our method outperforms well in classification performance and generalization. Particularly, we identified some significant subregions and genes, such as hippocampus amygdala transition area (HATA), fimbria, parasubiculum and genes included RYR3 and PRKCE. These discoveries provide some new candidate genes for AD and demonstrate the contribution of hippocampal subregions and genes to AD.
Keywords: clustering evolution; genetic algorithm; hippocampus; random forest; subregion.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Detection of Association Features Based on Gene Eigenvalues and MRI Imaging Using Genetic Weighted Random Forest.Genes (Basel). 2022 Dec 12;13(12):2344. doi: 10.3390/genes13122344. Genes (Basel). 2022. PMID: 36553611 Free PMC article.
-
Research on Pathogenic Hippocampal Voxel Detection in Alzheimer's Disease Using Clustering Genetic Random Forest.Front Psychiatry. 2022 Apr 7;13:861258. doi: 10.3389/fpsyt.2022.861258. eCollection 2022. Front Psychiatry. 2022. PMID: 35463515 Free PMC article.
-
Multimodal Data Analysis of Alzheimer's Disease Based on Clustering Evolutionary Random Forest.IEEE J Biomed Health Inform. 2020 Oct;24(10):2973-2983. doi: 10.1109/JBHI.2020.2973324. Epub 2020 Feb 11. IEEE J Biomed Health Inform. 2020. PMID: 32071013
-
A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer's disease.Neuroimage. 2020 Mar;208:116459. doi: 10.1016/j.neuroimage.2019.116459. Epub 2019 Dec 16. Neuroimage. 2020. PMID: 31837471
-
Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject-specific atlases.Neuroimage. 2016 Nov 1;141:542-555. doi: 10.1016/j.neuroimage.2016.07.020. Epub 2016 Jul 15. Neuroimage. 2016. PMID: 27426838 Free PMC article.
Cited by
-
Atrophy in subcortical gray matter in adult patients with moyamoya disease.Neurol Sci. 2023 May;44(5):1709-1717. doi: 10.1007/s10072-022-06583-x. Epub 2023 Jan 9. Neurol Sci. 2023. PMID: 36622475 Free PMC article.
-
Feature Fusion and Detection in Alzheimer's Disease Using a Novel Genetic Multi-Kernel SVM Based on MRI Imaging and Gene Data.Genes (Basel). 2022 May 7;13(5):837. doi: 10.3390/genes13050837. Genes (Basel). 2022. PMID: 35627222 Free PMC article.
-
miR-137: A therapeutic candidate or a key molecular regulator in Alzheimer's disease?J Alzheimers Dis Rep. 2025 Jun 25;9:25424823251352166. doi: 10.1177/25424823251352166. eCollection 2025 Jan-Dec. J Alzheimers Dis Rep. 2025. PMID: 40575117 Free PMC article. Review.
-
Detection of Association Features Based on Gene Eigenvalues and MRI Imaging Using Genetic Weighted Random Forest.Genes (Basel). 2022 Dec 12;13(12):2344. doi: 10.3390/genes13122344. Genes (Basel). 2022. PMID: 36553611 Free PMC article.
References
-
- Iglesias J.E., Augustinack J.C., Nguyen K., Player C.M., Player A., Wright M., Roy N., Frosch M.P., Mckee A.C., Wald L.L. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution mri: Application to adaptive segmentation of in vivo mri. Neuroimage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. - DOI - PMC - PubMed
-
- Cong S., Risacher S.L., West J.D., Wu Y.C., Apostolova L.G., Tallman E., Rizkalla M., Salama P., Saykin A.J., Shen L. Volumetric Comparison of Hippocampal Subfields Extracted from 4-Minute Accelerated versus 8-Minute High-resolution T2-weighted 3T MRI Scans. Brain Imaging Behav. 2018;12:1583–1595. doi: 10.1007/s11682-017-9819-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

