Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates
- PMID: 34062882
- PMCID: PMC8147356
- DOI: 10.3390/microorganisms9050984
Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates
Abstract
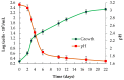
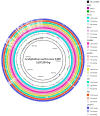
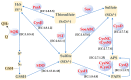
Acidiphilium multivorum LMS is an acidophile isolated from industrial bioreactors during the processing of the gold-bearing pyrite-arsenopyrite concentrate at 38-42 °C. Most strains of this species are obligate organoheterotrophs that do not use ferrous iron or reduced sulfur compounds as energy sources. However, the LMS strain was identified as one of the predominant sulfur oxidizers in acidophilic microbial consortia. In addition to efficient growth under strictly heterotrophic conditions, the LMS strain proved to be an active sulfur oxidizer both in the presence or absence of organic compounds. Interestingly, Ac. multivorum LMS was able to succeed more common sulfur oxidizers in microbial populations, which indicated a previously underestimated role of this bacterium in industrial bioleaching operations. In this study, the first draft genome of the sulfur-oxidizing Ac. multivorum was sequenced and annotated. Based on the functional genome characterization, sulfur metabolism pathways were reconstructed. The LMS strain possessed a complicated multi-enzyme system to oxidize elemental sulfur, thiosulfate, sulfide, and sulfite to sulfate as the final product. Altogether, the phenotypic description and genome analysis unraveled a crucial role of Ac. multivorum in some biomining processes and revealed unique strain-specific characteristics, including the ars genes conferring arsenic resistance, which are similar to those of phylogenetically distinct microorganisms.
Keywords: Acidiphilium multivorum; acidophilic microbial communities; arsenic resistance; biooxidation; gold-bearing sulfide concentrates; sulfur metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Harrison A.P., Jr. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int. J. Syst. Evol. Microbiol. 1981;31:327–332. doi: 10.1099/00207713-31-3-327. - DOI
-
- Wakao N., Nagasawa N., Matsuura T., Matsukura H., Matsumoto T., Hiraishi A., Matsumoto T., Hiraishi A., Sakurai Y., Shiota H. An acidophilic chemoorganotrophic bacterium from pyritic acid mine drainage. J. Gen. Appl. Microbiol. 1994;40:143–159. doi: 10.2323/jgam.40.143. - DOI
-
- Hiraishi A., Nagashima K.V.P., Matsuura K., Shimada K., Takaichi S., Wakao N., Katayama Y. Phylogeny and photosynthetic features of Thiobacillus acidophilus and related acidophilic bacteria: Its transfer to the genus Acidiphilium as Acidiphilium acidophilum comb. nov. Int. J. Syst. Bacteriol. 1998;48:1389–1398. doi: 10.1099/00207713-48-4-1389. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

