GeromiRs Are Downregulated in the Tumor Microenvironment during Colon Cancer Colonization of the Liver in a Murine Metastasis Model
- PMID: 34062897
- PMCID: PMC8124834
- DOI: 10.3390/ijms22094819
GeromiRs Are Downregulated in the Tumor Microenvironment during Colon Cancer Colonization of the Liver in a Murine Metastasis Model
Abstract
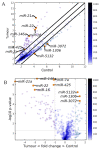
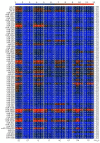
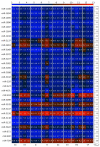
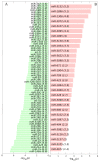
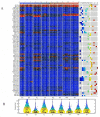
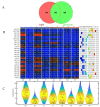
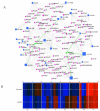
Cancer is a phenomenon broadly related to ageing in various ways such as cell cycle deregulation, metabolic defects or telomerases dysfunction as principal processes. Although the tumor cell is the main actor in cancer progression, it is not the only element of the disease. Cells and the matrix surrounding the tumor, called the tumor microenvironment (TME), play key roles in cancer progression. Phenotypic changes of the TME are indispensable for disease progression and a few of these transformations are produced by epigenetic changes including miRNA dysregulation. In this study, we found that a specific group of miRNAs in the liver TME produced by colon cancer called geromiRs, which are miRNAs related to the ageing process, are significantly downregulated. The three principal cell types involved in the liver TME, namely, liver sinusoidal endothelial cells, hepatic stellate (Ito) cells and Kupffer cells, were isolated from a murine hepatic metastasis model, and the miRNA and gene expression profiles were studied. From the 115 geromiRs and their associated hallmarks of aging, which we compiled from the literature, 75 were represented in the used microarrays, 26 out of them were downregulated in the TME cells during colon cancer colonization of the liver, and none of them were upregulated. The histone modification hallmark of the downregulated geromiRs is significantly enriched with the geromiRs miR-15a, miR-16, miR-26a, miR-29a, miR-29b and miR-29c. We built a network of all of the geromiRs downregulated in the TME cells and their gene targets from the MirTarBase database, and we analyzed the expression of these geromiR gene targets in the TME. We found that Cercam and Spsb4, identified as prognostic markers in a few cancer types, are associated with downregulated geromiRs and are upregulated in the TME cells.
Keywords: colorectal cancer; geromiRs; histone modifications; liver metastasis; miRNA; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Targeting liver sinusoidal endothelial cells with miR-20a-loaded nanoparticles reduces murine colon cancer metastasis to the liver.Int J Cancer. 2018 Aug 1;143(3):709-719. doi: 10.1002/ijc.31343. Epub 2018 Mar 15. Int J Cancer. 2018. PMID: 29492958
-
An Integrative Omics Approach Reveals Involvement of BRCA1 in Hepatic Metastatic Progression of Colorectal Cancer.Cancers (Basel). 2020 Aug 22;12(9):2380. doi: 10.3390/cancers12092380. Cancers (Basel). 2020. PMID: 32842712 Free PMC article.
-
The functional role of microRNAs in alcoholic liver injury.J Cell Mol Med. 2014 Feb;18(2):197-207. doi: 10.1111/jcmm.12223. Epub 2014 Jan 8. J Cell Mol Med. 2014. PMID: 24400890 Free PMC article. Review.
-
Interaction of non‑parenchymal hepatocytes in the process of hepatic fibrosis (Review).Mol Med Rep. 2021 May;23(5):364. doi: 10.3892/mmr.2021.12003. Epub 2021 Mar 24. Mol Med Rep. 2021. PMID: 33760176 Free PMC article. Review.
-
Systemic Delivery of scAAV8-Encoded MiR-29a Ameliorates Hepatic Fibrosis in Carbon Tetrachloride-Treated Mice.PLoS One. 2015 Apr 29;10(4):e0124411. doi: 10.1371/journal.pone.0124411. eCollection 2015. PLoS One. 2015. PMID: 25923107 Free PMC article.
Cited by
-
miRNAs in Cancer (Review of Literature).Int J Mol Sci. 2022 Mar 3;23(5):2805. doi: 10.3390/ijms23052805. Int J Mol Sci. 2022. PMID: 35269947 Free PMC article. Review.
-
CERCAM is a prognostic biomarker associated with immune infiltration of macrophage M2 polarization in head and neck squamous carcinoma.BMC Oral Health. 2023 Oct 7;23(1):724. doi: 10.1186/s12903-023-03421-0. BMC Oral Health. 2023. PMID: 37803318 Free PMC article.
-
Biological Roles and Clinical Therapeutic Applications of Tumor-Associated Macrophages in Colorectal Liver Metastasis.J Inflamm Res. 2024 Nov 7;17:8429-8443. doi: 10.2147/JIR.S493656. eCollection 2024. J Inflamm Res. 2024. PMID: 39529996 Free PMC article. Review.
-
Establishment of a tumor-associated fibroblast associated gene score based on scRNA-seq to predict prognosis in patients with triple-negative breast cancer.PLoS One. 2024 Oct 17;19(10):e0311801. doi: 10.1371/journal.pone.0311801. eCollection 2024. PLoS One. 2024. PMID: 39418248 Free PMC article.
-
Multi-omic analysis reveals metabolic pathways that characterize right-sided colon cancer liver metastasis.Cancer Lett. 2023 Oct 10;574:216384. doi: 10.1016/j.canlet.2023.216384. Epub 2023 Sep 14. Cancer Lett. 2023. PMID: 37716465 Free PMC article.
References
-
- American Cancer Society . Key Statistics for Colorectal Cancer. American Cancer Society; Atlanta, GA, USA: 2021.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

