HOXB7 Overexpression Leads Triple-Negative Breast Cancer Cells to a Less Aggressive Phenotype
- PMID: 34063128
- PMCID: PMC8148148
- DOI: 10.3390/biomedicines9050515
HOXB7 Overexpression Leads Triple-Negative Breast Cancer Cells to a Less Aggressive Phenotype
Abstract
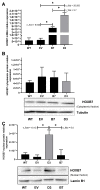
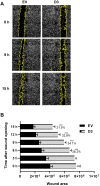
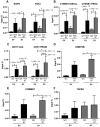
HOX genes appear to play a role in breast cancer progression in a molecular subtype-dependent way. The altered expression of HOXB7, for example, was reported to promote breast cancer progression in specific subtypes. Here we induced HOXB7 overexpression in MDA-MB-231 cells, a cellular model of the Triple-Negative breast cancer molecular subtype, and evaluated the phenotypic changes in cell viability, morphogenesis, migration, invasion, and colony formation. During the phenotypic characterization of the HOXB7-overexpressing cells, we consistently found less aggressive behavior represented by lower cell viability, inhibition of cell migration, invasion, and attachment-independent colony formation capacities added to the more compact and organized spheroid growth in 3D cultures. We then evaluated the expression of putative downstream targets and their direct binding to HOXB7 comparing ChIP-qPCR data generated from HOXB7-overexpressing cells and controls. In the manipulated cells, we found enriched biding of HOXB7 to CTNNB1, EGFR, FGF2, CDH1, DNMT3B, TGFB2, and COMMD7. Taken together, these results highlight the plasticity of the HOXB7 function in breast cancer, according to the cellular genetic background and expression levels, and provide evidence that in Triple-Negative breast cancer cells, HOXB7 overexpression has the potential to promote less aggressive phenotypes.
Keywords: HOXB7; MDA-MB-231; breast cancer; cell phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Epigenetic Regulation of CDH1 Is Altered after HOXB7-Silencing in MDA-MB-468 Triple-Negative Breast Cancer Cells.Genes (Basel). 2021 Oct 3;12(10):1575. doi: 10.3390/genes12101575. Genes (Basel). 2021. PMID: 34680970 Free PMC article.
-
HOXB7 promotes malignant progression by activating the TGFβ signaling pathway.Cancer Res. 2015 Feb 15;75(4):709-19. doi: 10.1158/0008-5472.CAN-14-3100. Epub 2014 Dec 26. Cancer Res. 2015. PMID: 25542862 Free PMC article.
-
Dataset of HOXB7, HOXB8 and HOXB9 expression profiles in cell lines representative of the breast cancer molecular subtypes Luminal a (MCF7), Luminal b (BT474), HER2+ (SKBR3) and triple-negative (MDA231, MDA468), compared to a model of normal cells (MCF10A).Data Brief. 2020 Apr 18;30:105572. doi: 10.1016/j.dib.2020.105572. eCollection 2020 Jun. Data Brief. 2020. PMID: 32346580 Free PMC article.
-
Calycosin inhibits triple-negative breast cancer progression through down-regulation of the novel estrogen receptor-α splice variant ER-α30-mediated PI3K/AKT signaling pathway.Phytomedicine. 2023 Sep;118:154924. doi: 10.1016/j.phymed.2023.154924. Epub 2023 Jun 14. Phytomedicine. 2023. PMID: 37393829
-
HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition.Cancer Res. 2006 Oct 1;66(19):9527-34. doi: 10.1158/0008-5472.CAN-05-4470. Cancer Res. 2006. PMID: 17018609
Cited by
-
Circ_0068631 sponges miR-139-5p to promote the growth and metastasis of cutaneous squamous cell carcinoma by upregulating HOXB7.Skin Res Technol. 2023 Feb;29(2):e13248. doi: 10.1111/srt.13248. Skin Res Technol. 2023. PMID: 36823512 Free PMC article.
-
Identification of Novel Prognostic Markers Associated With Laryngeal Squamous Cell Carcinoma Using Comprehensive Analysis.Front Oncol. 2022 Jan 11;11:779153. doi: 10.3389/fonc.2021.779153. eCollection 2021. Front Oncol. 2022. PMID: 35087752 Free PMC article.
-
Endorsement of TNBC Biomarkers in Precision Therapy by Nanotechnology.Cancers (Basel). 2023 May 8;15(9):2661. doi: 10.3390/cancers15092661. Cancers (Basel). 2023. PMID: 37174125 Free PMC article. Review.
-
Epigenetic Regulation of CDH1 Is Altered after HOXB7-Silencing in MDA-MB-468 Triple-Negative Breast Cancer Cells.Genes (Basel). 2021 Oct 3;12(10):1575. doi: 10.3390/genes12101575. Genes (Basel). 2021. PMID: 34680970 Free PMC article.
-
YAP1 activates SLC2A1 transcription and augments the malignant behavior of colorectal cancer cells by activating the Wnt/β-catenin signaling pathway.Cell Div. 2025 Apr 4;20(1):8. doi: 10.1186/s13008-025-00148-y. Cell Div. 2025. PMID: 40186232 Free PMC article.
References
-
- Li G., Hu J., Hu G. Biomarker Studies in Early Detection and Prognosis of Breast Cancer. In: Song E., Hu H., editors. Translational Research in Breast Cancer: Biomarker Diagnosis, Targeted Therapies and Approaches to Precision Medicine. Springer; Singapore: 2017. pp. 27–39. - PubMed
Grants and funding
- POCI-01-0145-FEDER-030562/FEDER - Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 - Operacional Programme for Competitiveness and Internationalisation
- PTDC/BTM-TEC/30562/2017/Portugal 2020, and Portuguese funds through FCT - Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

