Adenovirus Structure: What Is New?
- PMID: 34063479
- PMCID: PMC8156859
- DOI: 10.3390/ijms22105240
Adenovirus Structure: What Is New?
Abstract
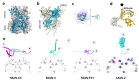
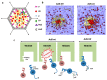
Adenoviruses are large (~950 Å) and complex non-enveloped, dsDNA icosahedral viruses. They have a pseudo-T = 25 triangulation number with at least 12 different proteins composing the virion. These include the major and minor capsid proteins, core proteins, maturation protease, terminal protein, and packaging machinery. Although adenoviruses have been studied for more than 60 years, deciphering their architecture has presented a challenge for structural biology techniques. An outstanding event was the first near-atomic resolution structure of human adenovirus type 5 (HAdV-C5), solved by cryo-electron microscopy (cryo-EM) in 2010. Discovery of new adenovirus types, together with methodological advances in structural biology techniques, in particular cryo-EM, has lately produced a considerable amount of new, high-resolution data on the organization of adenoviruses belonging to different species. In spite of these advances, the organization of the non-icosahedral core is still a great unknown. Nevertheless, alternative techniques such as atomic force microscopy (AFM) are providing interesting glimpses on the role of the core proteins in genome condensation and virion stability. Here we summarize the current knowledge on adenovirus structure, with an emphasis on high-resolution structures obtained since 2010.
Keywords: adenovirus; core proteins; cryo-EM; cryo-electron microscopy; crystallography; maturation; minor coat proteins; structure.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Berk A.J. Adenoviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Volume 1. Wolters Kluwer/Lippincott Williams & Wilkins Health; Philadelphia, PA, USA: 2013. pp. 1704–1731.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

