An Integrative Bioinformatic Analysis for Keratinase Detection in Marine-Derived Streptomyces
- PMID: 34063876
- PMCID: PMC8224001
- DOI: 10.3390/md19060286
An Integrative Bioinformatic Analysis for Keratinase Detection in Marine-Derived Streptomyces
Abstract
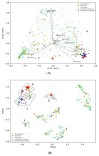
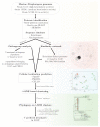
Keratinases present promising biotechnological applications, due to their ability to degrade keratin. Streptomyces appears as one of the main sources of these enzymes, but complete genome sequences of keratinolytic bacteria are still limited. This article reports the complete genomes of three marine-derived streptomycetes that show different levels of feather keratin degradation, with high (strain G11C), low (strain CHD11), and no (strain Vc74B-19) keratinolytic activity. A multi-step bioinformatics approach is described to explore genes encoding putative keratinases in these genomes. Despite their differential keratinolytic activity, multiplatform annotation reveals similar quantities of ORFs encoding putative proteases in strains G11C, CHD11, and Vc74B-19. Comparative genomics classified these putative proteases into 140 orthologous groups and 17 unassigned orthogroup peptidases belonging to strain G11C. Similarity network analysis revealed three network communities of putative peptidases related to known keratinases of the peptidase families S01, S08, and M04. When combined with the prediction of cellular localization and phylogenetic reconstruction, seven putative keratinases from the highly keratinolytic strain Streptomyces sp. G11C are identified. To our knowledge, this is the first multi-step bioinformatics analysis that complements comparative genomics with phylogeny and cellular localization prediction, for the prediction of genes encoding putative keratinases in streptomycetes.
Keywords: genomic comparison; keratinases; keratinolytic proteases; marine-derived Streptomyces.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C.Mar Drugs. 2020 Oct 28;18(11):537. doi: 10.3390/md18110537. Mar Drugs. 2020. PMID: 33126528 Free PMC article.
-
Bioinformatics based discovery of new keratinases in protease family M36.N Biotechnol. 2022 May 25;68:19-27. doi: 10.1016/j.nbt.2022.01.004. Epub 2022 Jan 12. N Biotechnol. 2022. PMID: 35032710
-
The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles.Sci Rep. 2017 Jul 5;7(1):4658. doi: 10.1038/s41598-017-04723-4. Sci Rep. 2017. PMID: 28680127 Free PMC article.
-
Comprehensive insights into microbial keratinases and their implication in various biotechnological and industrial sectors: A review.Int J Biol Macromol. 2020 Jul 1;154:567-583. doi: 10.1016/j.ijbiomac.2020.03.116. Epub 2020 Mar 16. Int J Biol Macromol. 2020. PMID: 32194110 Review.
-
Microbial enzymes catalyzing keratin degradation: Classification, structure, function.Biotechnol Adv. 2020 Nov 15;44:107607. doi: 10.1016/j.biotechadv.2020.107607. Epub 2020 Aug 5. Biotechnol Adv. 2020. PMID: 32768519 Free PMC article. Review.
Cited by
-
Enzyme Bioprospection of Marine-Derived Actinobacteria from the Chilean Coast and New Insight in the Mechanism of Keratin Degradation in Streptomyces sp. G11C.Mar Drugs. 2020 Oct 28;18(11):537. doi: 10.3390/md18110537. Mar Drugs. 2020. PMID: 33126528 Free PMC article.
-
An update on thermostable keratinases for protein engineering against feather pollutants.Appl Microbiol Biotechnol. 2025 Mar 25;109(1):75. doi: 10.1007/s00253-025-13459-5. Appl Microbiol Biotechnol. 2025. PMID: 40131452 Free PMC article. Review.
-
Description and Genomic Characterization of Oceaniferula flavus sp. nov., a Novel Potential Polysaccharide-Degrading Candidate of the Difficult-to-Cultivate Phylum Verrucomicrobiota Isolated from Seaweed.Mar Drugs. 2022 Dec 29;21(1):31. doi: 10.3390/md21010031. Mar Drugs. 2022. PMID: 36662204 Free PMC article.
References
-
- Brandelli A. Bacterial keratinases: Useful enzymes for bioprocessing agroindustrial wastes and beyond. Food Bioprocess Technol. 2008;1:105–116. doi: 10.1007/s11947-007-0025-y. - DOI
MeSH terms
Substances
Grants and funding
- Fondecyt 1171555/Comisión Nacional de Investigación Científica y Tecnológica
- CONICYT PIA ACT172128/Comisión Nacional de Investigación Científica y Tecnológica
- PhD fellowship 21161188/Comisión Nacional de Investigación Científica y Tecnológica
- POSTDOCTORADO 3180399/Comisión Nacional de Investigación Científica y Tecnológica
- PhD fellowship N° 21180908/Comisión Nacional de Investigación Científica y Tecnológica
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

