Molecular Insights into the Genetic Variability of ORF Virus in a Mediterranean Region (Sardinia, Italy)
- PMID: 34064326
- PMCID: PMC8147818
- DOI: 10.3390/life11050416
Molecular Insights into the Genetic Variability of ORF Virus in a Mediterranean Region (Sardinia, Italy)
Abstract
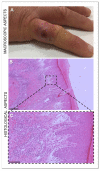
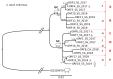
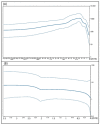
Orf virus (ORFV) represents the causative agent of contagious ecthyma, clinically characterized by mild papular and pustular to severe proliferative lesions, mainly occurring in sheep and goats. In order to provide hints on the evolutionary history of this virus, we carried out a study aimed to assess the genetic variation of ORFV in Sardinia that hosts a large affected small ruminant population. We also found a high worldwide mutational viral evolutionary rate, which resulted, in turn, higher than the rate we detected for the strains isolated in Sardinia. In addition, a well-supported genetic divergence was found between the viral strains isolated from sheep and those from goats, but no relevant connection was evidenced between the severity of lesions produced by ORFV and specific polymorphic patterns in the two species of hosts. Such a finding suggests that ORFV infection-related lesions are not necessarily linked to the expression of one of the three genes here analyzed and could rather be the effect of the expression of other genes or rather represents a multifactorial character.
Keywords: B2L; O45; VIR; contagious ecthyma; molecular dating; phylogenetic analysis; sequencing.
Conflict of interest statement
The authors and the founders declare no conflict of interest.
Figures




References
-
- Vaccari F. Ph.D. Thesis. University of Bologna; Bologna, Italy: 2009. Evolutionary Mechanisms of Parapoxviruses: Genomic Characterization of Pseudocowpoxviruses and Development of Systems for the Study of Recombinations.
-
- Zeller H. South-West African Goat Pox. Arb. Reichsgesundheitsamte. 1920;52:501–537.
-
- Kumar R., Trivedi R.N., Bhatt P., Khan S.H., Khurana S.K., Tiwari R., Karthik K., Malik Y.S., Dhama K., Chandra R. Contagious pustular dermatitis (orf disease)—Epidemiology, diagnosis, control and public health concerns. Adv. Anim. Vet. Sci. 2015;3:649–676. doi: 10.14737/journal.aavs/2015/3.12.649.676. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

