From Structural Studies to HCV Vaccine Design
- PMID: 34064532
- PMCID: PMC8147963
- DOI: 10.3390/v13050833
From Structural Studies to HCV Vaccine Design
Abstract
Hepatitis C virus (HCV) is a serious and growing public health problem despite recent developments of antiviral therapeutics. To achieve global elimination of HCV, an effective cross-genotype vaccine is needed. The failure of previous vaccination trials to elicit an effective cross-reactive immune response demands better vaccine antigens to induce a potent cross-neutralizing response to improve vaccine efficacy. HCV E1 and E2 envelope (Env) glycoproteins are the main targets for neutralizing antibodies (nAbs), which aid in HCV clearance and protection. Therefore, a molecular-level understanding of the nAb responses against HCV is imperative for the rational design of cross-genotype vaccine antigens. Here we summarize the recent advances in structural studies of HCV Env and Env-nAb complexes and how they improve our understanding of immune recognition of HCV. We review the structural data defining HCV neutralization epitopes and conformational plasticity of the Env proteins, and the knowledge applicable to rational vaccine design.
Keywords: E1; E1E2 complex; E2; VH1-69; envelope glycoproteins; hepatitis C virus (HCV); neutralization face; neutralizing antibodies; structural studies; vaccine design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
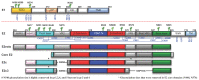
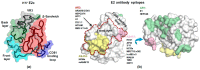
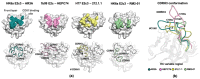

References
-
- New Hepatitis Data Highlight Need for Urgent Global Response. [(accessed on 27 April 2021)];2017 Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
-
- Petruzziello A., Marigliano S., Loquercio G., Cozzolino A., Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J. Gastroenterol. 2016;22:7824–7840. doi: 10.3748/wjg.v22.i34.7824. - DOI - PMC - PubMed
-
- Surveillance for Viral Hepatitis—United States 2017. [(accessed on 27 April 2021)];2019 Available online: https://www.cdc.gov/hepatitis/statistics/2017surveillance/index.htm.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

