A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease
- PMID: 34064787
- PMCID: PMC8150904
- DOI: 10.3390/cells10051164
A Biochemical and Structural Understanding of TOM Complex Interactions and Implications for Human Health and Disease
Abstract
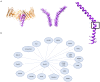
The central role mitochondria play in cellular homeostasis has made its study critical to our understanding of various aspects of human health and disease. Mitochondria rely on the translocase of the outer membrane (TOM) complex for the bulk of mitochondrial protein import. In addition to its role as the major entry point for mitochondrial proteins, the TOM complex serves as an entry pathway for viral proteins. TOM complex subunits also participate in a host of interactions that have been studied extensively for their function in neurodegenerative diseases, cardiovascular diseases, innate immunity, cancer, metabolism, mitophagy and autophagy. Recent advances in our structural understanding of the TOM complex and the protein import machinery of the outer mitochondrial membrane have made structure-based therapeutics targeting outer mitochondrial membrane proteins during mitochondrial dysfunction an exciting prospect. Here, we describe advances in understanding the TOM complex, the interactome of the TOM complex subunits, the implications for the development of therapeutics, and our understanding of the structure/function relationship between components of the TOM complex and mitochondrial homeostasis.
Keywords: TOM complex; TOM complex interactions; TOM subunits; mitochondrial cell signaling; mitochondrial quality control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

