Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population
- PMID: 34064921
- PMCID: PMC8150974
- DOI: 10.3390/genes12050719
Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population
Abstract
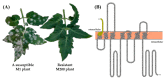
In tomato (Solanum lycopersicum), there are at least three SlMLO (Mildew resistance Locus O) genes acting as susceptibility genes for the powdery mildew disease caused by Oidium neolycopersici, namely SlMLO1, SlMLO5 and SlMLO8. Of the three homologs, the SlMLO1 gene plays a major role since a natural mutant allele called ol-2 can almost completely prevent fungal penetration by formation of papillae. The ol-2 allele contains a 19-bp deletion in the coding sequence of the SlMLO1 gene, resulting in a premature stop codon within the second cytoplasmic loop of the predicted protein. In this study, we have developed a new genetic resource (M200) in the tomato cv. Micro-Tom genetic background by means of ethyl methane sulfonate (EMS) mutagenesis. The mutant M200 containing a novel allele (the m200 allele) of the tomato SlMLO1 gene showed profound resistance against powdery mildew with no fungal sporulation. Compared to the coding sequence of the SlMLO1 gene, the m200 allele carries a point mutation at T65A. The SNP results in a premature stop codon L22* located in the first transmembrane domain of the complete SlMLO1 protein. The length of the predicted protein is 21 amino acids, while the SlMLO1 full-length protein is 513 amino acids. A high-resolution melting (HRM) marker was developed to distinguish the mutated m200 allele from the SlMLO1 allele in backcross populations. The mutant allele conferred recessive resistance that was associated with papillae formation at fungal penetration sites of plant epidermal cells. A comprehensive list of known mlo mutations found in natural and artificial mutants is presented, which serves as a particularly valuable resource for powdery mildew resistance breeding.
Keywords: EMS mutagenesis; Micro-Tom; Oidium neolycopersici; SlMLO1; Solanum lycopersicum; powdery mildew.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Development of DNA markers for Slmlo1.1, a new mutant allele of the powdery mildew resistance gene SlMlo1 in tomato (Solanum lycopersicum).Genome. 2018 Oct;61(10):703-712. doi: 10.1139/gen-2018-0114. Epub 2018 Aug 22. Genome. 2018. PMID: 30134125
-
Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica.PLoS One. 2013 Jul 29;8(7):e70723. doi: 10.1371/journal.pone.0070723. Print 2013. PLoS One. 2013. PMID: 23923019 Free PMC article.
-
Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function.Mol Plant Microbe Interact. 2008 Jan;21(1):30-9. doi: 10.1094/MPMI-21-1-0030. Mol Plant Microbe Interact. 2008. PMID: 18052880
-
Magical mystery tour: MLO proteins in plant immunity and beyond.New Phytol. 2014 Oct;204(2):273-81. doi: 10.1111/nph.12889. New Phytol. 2014. PMID: 25453131 Review.
-
Serpentine plant MLO proteins as entry portals for powdery mildew fungi.Biochem Soc Trans. 2005 Apr;33(Pt 2):389-92. doi: 10.1042/BST0330389. Biochem Soc Trans. 2005. PMID: 15787613 Review.
Cited by
-
Less is more: CRISPR/Cas9-based mutations in DND1 gene enhance tomato resistance to powdery mildew with low fitness costs.BMC Plant Biol. 2024 Aug 10;24(1):763. doi: 10.1186/s12870-024-05428-3. BMC Plant Biol. 2024. PMID: 39123110 Free PMC article.
-
MLO Proteins from Tomato (Solanum lycopersicum L.) and Related Species in the Broad Phylogenetic Context.Plants (Basel). 2022 Jun 16;11(12):1588. doi: 10.3390/plants11121588. Plants (Basel). 2022. PMID: 35736740 Free PMC article.
-
Mutation of PUB21 in tomato leads to reduced susceptibility to necrotrophic fungi.BMC Plant Biol. 2025 Aug 8;25(1):1038. doi: 10.1186/s12870-025-07107-3. BMC Plant Biol. 2025. PMID: 40781591 Free PMC article.
-
Germplasm Screening Using DNA Markers and Genome-Wide Association Study for the Identification of Powdery Mildew Resistance Loci in Tomato.Int J Mol Sci. 2022 Nov 6;23(21):13610. doi: 10.3390/ijms232113610. Int J Mol Sci. 2022. PMID: 36362397 Free PMC article.
-
Current trends and insights on EMS mutagenesis application to studies on plant abiotic stress tolerance and development.Front Plant Sci. 2023 Jan 4;13:1052569. doi: 10.3389/fpls.2022.1052569. eCollection 2022. Front Plant Sci. 2023. PMID: 36684716 Free PMC article. Review.
References
-
- Schijlen E.G., de Vos C.R., Martens S., Jonker H.H., Rosin F.M., Molthoff J.W., Tikunov Y.M., Angenent G.C., van Tunen A.J., Bovy A.G. RNA Interference Silencing of Chalcone Synthase, the First Step in the Flavonoid Biosynthesis Pathway, Leads to Parthenocarpic Tomato Fruits. Plant. Physiol. 2007;144:1520–1530. doi: 10.1104/pp.107.100305. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

