Retrospective Data Insight into the Global Distribution of Carbapenemase-Producing Pseudomonas aeruginosa
- PMID: 34065054
- PMCID: PMC8151531
- DOI: 10.3390/antibiotics10050548
Retrospective Data Insight into the Global Distribution of Carbapenemase-Producing Pseudomonas aeruginosa
Abstract
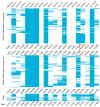
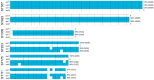
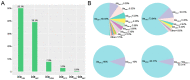
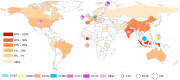
This study aimed to determine the global distribution and molecular characteristics of carbapenemase-producing Pseudomonas aeruginosa isolates. A total of 328 (11.1%, 328/2953) carbapenemase-producing P. aeruginosa isolates from humans were obtained from public databases as of October 2019. Of which, the blaVIM and blaIMP genes were the most prevalent carbapenemases in the P. aeruginosa isolates. These carbapenemase-producing P. aeruginosa isolates possessed 34 distinct sequence types (STs) and six predominated: ST357, ST823, ST308, ST233, ST175 and ST111. The ST357 and ST823 isolates were primarily found detected in Asia and all ST175 isolates were found in Europe. The ST308, ST233 and ST111 isolates were spread worldwide. Further, all ST823 isolates and the majority of ST111, ST233 and ST175 isolates carried blaVIM but ST357 isolates primarily carried blaIMP. ST308 isolates provide a key reservoir for the spread of blaVIM, blaIMP and blaNDM. WGS analysis revealed that ST111 carried a great diversity of ARG types (n = 23), followed by ST357 (n = 21), ST308 (n = 19), ST233 (n = 18), ST175 (n = 14) and ST823 (n = 10). The ST175 isolates carried a more diversity and frequent of aminoglycoside ARGs, and ST233 isolates harbored more tetracycline ARGs. Our findings revealed that different carbapenem resistance genes were distributed primarily in variant STs of P. aeruginosa isolates, these isolates also possessed an extensive geographical distribution that highlights the need for surveillance studies that detect carbapenemase-producing P. aeruginosa isolates in humans.
Keywords: ARGs; MLST; P. aeruginosa; carbapenemase; global distribution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hawkey J., Le Hello S., Doublet B., Granier S.A., Hendriksen R.S., Fricke W.F., Ceyssens P.-J., Gomart C., Billman-Jacobe H., Holt K.E., et al. Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb. Genom. 2019;5:e000269. doi: 10.1099/mgen.0.000269. - DOI - PMC - PubMed
-
- Wyres K.L., Hawkey J., Hetland M.A.K., Fostervold A., Wick R.R., Judd L.M., Hamidian M., Howden B.P., Löhr I.H., Holt K.E. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J. Antimicrob. Chemother. 2019;74:577–581. doi: 10.1093/jac/dky492. - DOI - PMC - PubMed
-
- Jia H., Chen Y., Wang J., Xie X., Ruan Z. Emerging challenges of whole-genome-sequencing-powered epidemiological surveillance of globally distributed clonal groups of bacterial infections, giving Acinetobacter baumannii ST195 as an example. Int. J. Med. Microbiol. 2019;309:151339. doi: 10.1016/j.ijmm.2019.151339. - DOI - PubMed
Grants and funding
- 31802243/National Natural Science Foundation of China
- 31730097/National Natural Science Foundation of China
- 31772793/National Natural Science Foundation of China
- IRT_17R39/Program for Innovative Research Team in the University of Ministry of Education of China
- 2019BT02N054/Guangdong Special Support Program Innovation Team
LinkOut - more resources
Full Text Sources
Miscellaneous

