Tandem Repeats in Bacillus: Unique Features and Taxonomic Distribution
- PMID: 34065296
- PMCID: PMC8161180
- DOI: 10.3390/ijms22105373
Tandem Repeats in Bacillus: Unique Features and Taxonomic Distribution
Abstract
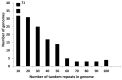
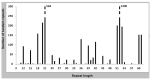
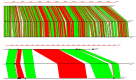
Little is known about DNA tandem repeats across prokaryotes. We have recently described an enigmatic group of tandem repeats in bacterial genomes with a constant repeat size but variable sequence. These findings strongly suggest that tandem repeat size in some bacteria is under strong selective constraints. Here, we extend these studies and describe tandem repeats in a large set of Bacillus. Some species have very few repeats, while other species have a large number. Most tandem repeats have repeats with a constant size (either 52 or 20-21 nt), but a variable sequence. We characterize in detail these intriguing tandem repeats. Individual species have several families of tandem repeats with the same repeat length and different sequence. This result is in strong contrast with eukaryotes, where tandem repeats of many sizes are found in any species. We discuss the possibility that they are transcribed as small RNA molecules. They may also be involved in the stabilization of the nucleoid through interaction with proteins. We also show that the distribution of tandem repeats in different species has a taxonomic significance. The data we present for all tandem repeats and their families in these bacterial species will be useful for further genomic studies.
Keywords: Bacillus; Bacillus coagulans; bacteria; bacterial nucleoid; non-coding DNA; satellites; small RNA; tandem repeats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

