Rapamycin Ameliorates Defects in Mitochondrial Fission and Mitophagy in Glioblastoma Cells
- PMID: 34065350
- PMCID: PMC8161366
- DOI: 10.3390/ijms22105379
Rapamycin Ameliorates Defects in Mitochondrial Fission and Mitophagy in Glioblastoma Cells
Abstract
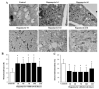
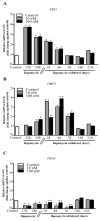
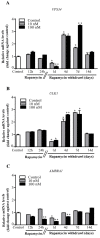
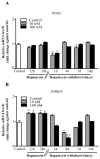
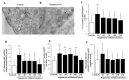
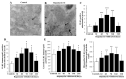
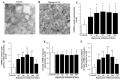
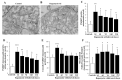
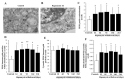
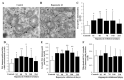
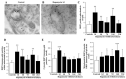
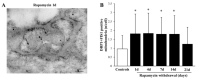
Glioblastoma (GBM) cells feature mitochondrial alterations, which are documented and quantified in the present study, by using ultrastructural morphometry. Mitochondrial impairment, which roughly occurs in half of the organelles, is shown to be related to mTOR overexpression and autophagy suppression. The novelty of the present study consists of detailing an mTOR-dependent mitophagy occlusion, along with suppression of mitochondrial fission. These phenomena contribute to explain the increase in altered mitochondria reported here. Administration of the mTOR inhibitor rapamycin rescues mitochondrial alterations. In detail, rapamycin induces the expression of genes promoting mitophagy (PINK1, PARKIN, ULK1, AMBRA1) and mitochondrial fission (FIS1, DRP1). This occurs along with over-expression of VPS34, an early gene placed upstream in the autophagy pathway. The topographic stoichiometry of proteins coded by these genes within mitochondria indicates that, a remarkable polarization of proteins involved in fission and mitophagy within mitochondria including LC3 takes place. Co-localization of these proteins within mitochondria, persists for weeks following rapamycin, which produces long-lasting mitochondrial plasticity. Thus, rapamycin restores mitochondrial status in GBM cells. These findings add novel evidence about mitochondria and GBM, while fostering a novel therapeutic approach to restore healthy mitochondria through mTOR inhibition.
Keywords: AMBRA1; DRP1; FIS1; OPA1; PARKIN; PINK1; ULK1; VPS34; autophagy; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Autophagy Activation Associates with Suppression of Prion Protein and Improved Mitochondrial Status in Glioblastoma Cells.Cells. 2023 Jan 4;12(2):221. doi: 10.3390/cells12020221. Cells. 2023. PMID: 36672156 Free PMC article.
-
Quantitative Ultrastructural Morphometry and Gene Expression of mTOR-Related Mitochondriogenesis within Glioblastoma Cells.Int J Mol Sci. 2020 Jun 27;21(13):4570. doi: 10.3390/ijms21134570. Int J Mol Sci. 2020. PMID: 32604996 Free PMC article.
-
Silibinin-induced mitochondria fission leads to mitophagy, which attenuates silibinin-induced apoptosis in MCF-7 and MDA-MB-231 cells.Arch Biochem Biophys. 2020 May 30;685:108284. doi: 10.1016/j.abb.2020.108284. Epub 2020 Jan 31. Arch Biochem Biophys. 2020. PMID: 32014401
-
Dynamin-related protein 1: A protein critical for mitochondrial fission, mitophagy, and neuronal death in Parkinson's disease.Pharmacol Res. 2020 Jan;151:104553. doi: 10.1016/j.phrs.2019.104553. Epub 2019 Nov 21. Pharmacol Res. 2020. PMID: 31760107 Review.
-
Mitochondrial dynamics in type 2 diabetes: Pathophysiological implications.Redox Biol. 2017 Apr;11:637-645. doi: 10.1016/j.redox.2017.01.013. Epub 2017 Jan 16. Redox Biol. 2017. PMID: 28131082 Free PMC article. Review.
Cited by
-
Connexin Expression in Human Minor Salivary Glands: An Immunohistochemical Microscopy Study.Molecules. 2022 Sep 12;27(18):5926. doi: 10.3390/molecules27185926. Molecules. 2022. PMID: 36144660 Free PMC article.
-
Molecular mechanisms of mitochondrial homeostasis regulation in neurons and possible therapeutic approaches for Alzheimer's disease.Heliyon. 2024 Aug 17;10(17):e36470. doi: 10.1016/j.heliyon.2024.e36470. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39281517 Free PMC article. Review.
-
Autophagy Activation Associates with Suppression of Prion Protein and Improved Mitochondrial Status in Glioblastoma Cells.Cells. 2023 Jan 4;12(2):221. doi: 10.3390/cells12020221. Cells. 2023. PMID: 36672156 Free PMC article.
-
Synaptic Alterations in a Transgenic Model of Tuberous Sclerosis Complex: Relevance to Autism Spectrum Disorders.Int J Mol Sci. 2021 Sep 17;22(18):10058. doi: 10.3390/ijms221810058. Int J Mol Sci. 2021. PMID: 34576223 Free PMC article.
-
Restoring the infected powerhouse: Mitochondrial quality control in sepsis.Redox Biol. 2023 Dec;68:102968. doi: 10.1016/j.redox.2023.102968. Epub 2023 Nov 23. Redox Biol. 2023. PMID: 38039825 Free PMC article. Review.
References
-
- Jiang H., White E.J., Conrad C., Gomez-Manzano C., Fueyo J. Autophagy pathways in glioblastoma. Methods Enzymol. 2009;453:273–286. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

