SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability
- PMID: 34065507
- PMCID: PMC8161396
- DOI: 10.3390/ijms22105391
SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability
Abstract
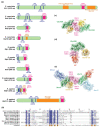
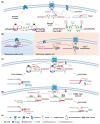
Small ubiquitin-like modifier (SUMO)-targeted E3 ubiquitin ligases (STUbLs) are specialized enzymes that recognize SUMOylated proteins and attach ubiquitin to them. They therefore connect the cellular SUMOylation and ubiquitination circuits. STUbLs participate in diverse molecular processes that span cell cycle regulated events, including DNA repair, replication, mitosis, and transcription. They operate during unperturbed conditions and in response to challenges, such as genotoxic stress. These E3 ubiquitin ligases modify their target substrates by catalyzing ubiquitin chains that form different linkages, resulting in proteolytic or non-proteolytic outcomes. Often, STUbLs function in compartmentalized environments, such as the nuclear envelope or kinetochore, and actively aid in nuclear relocalization of damaged DNA and stalled replication forks to promote DNA repair or fork restart. Furthermore, STUbLs reside in the same vicinity as SUMO proteases and deubiquitinases (DUBs), providing spatiotemporal control of their targets. In this review, we focus on the molecular mechanisms by which STUbLs help to maintain genome stability across different species.
Keywords: RNF111; RNF4; STUbL; SUMO; Slx5/Slx8; genome stability; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kotsantis P., Petermann E., Boulton S.J. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov. 2018;8:537–555. doi: 10.1158/2159-8290.CD-17-1461. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

