An Analytical Review of the Structural Features of Pentatricopeptide Repeats: Strategic Amino Acids, Repeat Arrangements and Superhelical Architecture
- PMID: 34065603
- PMCID: PMC8160929
- DOI: 10.3390/ijms22105407
An Analytical Review of the Structural Features of Pentatricopeptide Repeats: Strategic Amino Acids, Repeat Arrangements and Superhelical Architecture
Abstract
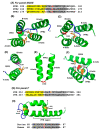
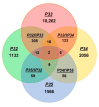
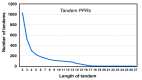
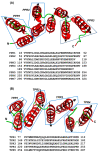
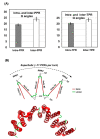
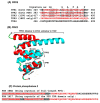
Tricopeptide repeats are common in natural proteins, and are exemplified by 34- and 35-residue repeats, known respectively as tetratricopeptide repeats (TPRs) and pentatricopeptide repeats (PPRs). In both classes, each repeat unit forms an antiparallel bihelical structure, so that multiple such units in a polypeptide are arranged in a parallel fashion. The primary structures of the motifs are nonidentical, but amino acids of similar properties occur in strategic positions. The focus of the present work was on PPR, but TPR, its better-studied cousin, is often included for comparison. The analyses revealed that critical amino acids, namely Gly, Pro, Ala and Trp, were placed at distinct locations in the higher order structure of PPR domains. While most TPRs occur in repeats of three, the PPRs exhibited a much greater diversity in repeat numbers, from 1 to 30 or more, separated by spacers of various sequences and lengths. Studies of PPR strings in proteins showed that the majority of PPR units are single, and that the longer tandems (i.e., without space in between) occurred in decreasing order. The multi-PPR domains also formed superhelical vortices, likely governed by interhelical angles rather than the spacers. These findings should be useful in designing and understanding the PPR domains.
Keywords: PPR; helix; protein structure; protein-RNA interaction; solvation; tricopeptide repeats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Nature and Arrangement of Pentatricopeptide Domains and the Linker Sequences Between Them.Bioinform Biol Insights. 2020 Mar 4;14:1177932220906434. doi: 10.1177/1177932220906434. eCollection 2020. Bioinform Biol Insights. 2020. PMID: 32180683 Free PMC article.
-
A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins.PLoS Genet. 2012;8(8):e1002910. doi: 10.1371/journal.pgen.1002910. Epub 2012 Aug 16. PLoS Genet. 2012. PMID: 22916040 Free PMC article.
-
Pentatricopeptide repeat proteins and their emerging roles in plants.Plant Physiol Biochem. 2007 Aug;45(8):521-34. doi: 10.1016/j.plaphy.2007.03.026. Epub 2007 Mar 24. Plant Physiol Biochem. 2007. PMID: 17560114 Review.
-
The design and structural characterization of a synthetic pentatricopeptide repeat protein.Acta Crystallogr D Biol Crystallogr. 2015 Feb;71(Pt 2):196-208. doi: 10.1107/S1399004714024869. Epub 2015 Jan 23. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25664731
-
[Advance in the study of PPR gene family].Yi Chuan. 2006 Jun;28(6):726-30. Yi Chuan. 2006. PMID: 16818438 Review. Chinese.
Cited by
-
Special Issue: Structure, Function and Evolution of Protein Domains.Int J Mol Sci. 2022 May 31;23(11):6201. doi: 10.3390/ijms23116201. Int J Mol Sci. 2022. PMID: 35682878 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

