Disruption of YY1-EZH2 Interaction Using Synthetic Peptides Inhibits Breast Cancer Development
- PMID: 34065631
- PMCID: PMC8156467
- DOI: 10.3390/cancers13102402
Disruption of YY1-EZH2 Interaction Using Synthetic Peptides Inhibits Breast Cancer Development
Abstract
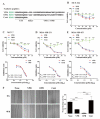
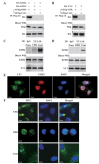
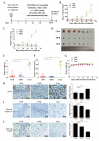
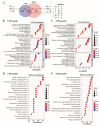
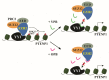
Enhancer of zeste homolog 2 (EZH2) is a methyltransferase to mediate lysine 27 trimethylation in histone H3 (i.e., H3K27me3) and repress gene expression. In solid tumors, EZH2 promotes oncogenesis and is considered a therapeutic target. As a transcription factor, Yin Yang 1 (YY1) recruits EZH2 through its oncoprotein binding (OPB) domain to establish gene repression. In this study, we mapped the YY1 protein binding (YPB) domain on EZH2 to a region of 27 amino acids. Both YPB and OPB domain synthetic peptides could disrupt YY1EZH2 interaction, markedly reduce breast cancer cell viability, and efficiently inhibit tumor growth in a xenograft mouse model. We analyzed MDA-MB-231 cells treated with YPB, OPB, and control peptides by chromatin immunoprecipitation DNA sequencing (ChIP-seq) using an antibody against H3K27me3. YPB and OPB treatments altered H3K27me3 on 465 and 1137 genes, respectively, compared to the control. Of these genes, 145 overlapped between the two peptides. Among them, PTENP1, the PTEN pseudogene, showed reduced H3K27me3 signal when treated by either YPB or OPB peptide. Consistently, the two peptides enhanced both PTENP1 and PTEN expression with concomitantly reduced AKT activation. Further studies validated PTENP1's contribution to the anticancer activity of YPB and OPB peptides.
Keywords: EZH2; OPB; PTENP1; YPB; YY1; breast cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

