Discovery of Orphan Olfactory Receptor 6M1 as a New Anticancer Target in MCF-7 Cells by a Combination of Surface Plasmon Resonance-Based and Cell-Based Systems
- PMID: 34065710
- PMCID: PMC8156394
- DOI: 10.3390/s21103468
Discovery of Orphan Olfactory Receptor 6M1 as a New Anticancer Target in MCF-7 Cells by a Combination of Surface Plasmon Resonance-Based and Cell-Based Systems
Abstract
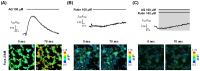
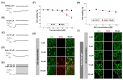
Olfactory receptors (ORs) account for 49% of all G protein-coupled receptors (GPCRs), which are important targets for drug discovery, and hence ORs may also be potential drug targets. Various ORs are expressed in breast cancer cells; however, most of them are orphan receptors, and thus, their functions are unknown. Herein, we present an experimental strategy using a surface plasmon resonance (SPR) system and a cell-based assay that allowed the identification of orphan OR6M1 as a new anticancer target in the MCF-7 breast cancer cell line. After the construction of stable OR6M1-expressing cells, the SPR-based screening of 108 chemicals for ligand activity was performed against OR6M1-expressing whole cells (primary screening) or membrane fragments (secondary screening). As a result, anthraquinone (AQ) and rutin were discovered to be new OR6M1 ligands. Based on calcium imaging in OR6M1-expressing Hana3A cells, AQ and rutin were classified as an OR6M1 agonist and antagonist, respectively. Cell viability and live/dead assays showed that AQ induced the death of MCF-7 cells, which was inhibited by rutin. Therefore, OR6M1 may be considered an anticancer target, and AQ may be considered a chemotherapeutic agent. This combined method can be widely used to discover the ligands and functions of other orphan GPCRs.
Keywords: MCF-7 breast cancer cell line; OR6M1; anthraquinone; olfactory receptor; rutin; surface plasmon resonance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Discovery of (-)-epigallocatechin gallate, a novel olfactory receptor 2AT4 agonist that regulates proliferation and apoptosis in leukemia cells.Heliyon. 2024 May 5;10(10):e30298. doi: 10.1016/j.heliyon.2024.e30298. eCollection 2024 May 30. Heliyon. 2024. PMID: 38778941 Free PMC article.
-
Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery.Biochim Biophys Acta. 2014 Jan;1838(1 Pt A):43-55. doi: 10.1016/j.bbamem.2013.04.028. Epub 2013 May 9. Biochim Biophys Acta. 2014. PMID: 23665295 Review.
-
Surface plasmon resonance analysis of seven-transmembrane receptors.Methods Enzymol. 2015;556:499-525. doi: 10.1016/bs.mie.2015.01.016. Epub 2015 Mar 21. Methods Enzymol. 2015. PMID: 25857797 Review.
-
Real-time monitoring of odorant-induced cellular reactions using surface plasmon resonance.Biosens Bioelectron. 2009 Sep 15;25(1):55-60. doi: 10.1016/j.bios.2009.06.007. Epub 2009 Jun 10. Biosens Bioelectron. 2009. PMID: 19559592
-
Detection of G protein-coupled receptor-mediated cellular response involved in cytoskeletal rearrangement using surface plasmon resonance.Biosens Bioelectron. 2010 Mar 15;25(7):1675-80. doi: 10.1016/j.bios.2009.12.006. Epub 2009 Dec 16. Biosens Bioelectron. 2010. PMID: 20044245
Cited by
-
Machine Learning Assisted Approach for Finding Novel High Activity Agonists of Human Ectopic Olfactory Receptors.Int J Mol Sci. 2021 Oct 26;22(21):11546. doi: 10.3390/ijms222111546. Int J Mol Sci. 2021. PMID: 34768977 Free PMC article.
-
Odorant receptors in cancer.BMB Rep. 2022 Feb;55(2):72-80. doi: 10.5483/BMBRep.2022.55.2.010. BMB Rep. 2022. PMID: 35168702 Free PMC article. Review.
-
Biosensors for Odor Detection: A Review.Biosensors (Basel). 2023 Nov 27;13(12):1000. doi: 10.3390/bios13121000. Biosensors (Basel). 2023. PMID: 38131760 Free PMC article. Review.
-
Discovery of (-)-epigallocatechin gallate, a novel olfactory receptor 2AT4 agonist that regulates proliferation and apoptosis in leukemia cells.Heliyon. 2024 May 5;10(10):e30298. doi: 10.1016/j.heliyon.2024.e30298. eCollection 2024 May 30. Heliyon. 2024. PMID: 38778941 Free PMC article.
-
Acute Myeloid Leukemia Expresses a Specific Group of Olfactory Receptors.Cancers (Basel). 2023 Jun 6;15(12):3073. doi: 10.3390/cancers15123073. Cancers (Basel). 2023. PMID: 37370684 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

