Pan-Genome miRNomics in Brachypodium
- PMID: 34065739
- PMCID: PMC8156279
- DOI: 10.3390/plants10050991
Pan-Genome miRNomics in Brachypodium
Abstract
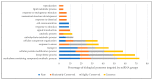
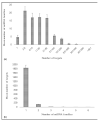
Pan-genomes are efficient tools for the identification of conserved and varying genomic sequences within lineages of a species. Investigating genetic variations might lead to the discovery of genes present in a subset of lineages, which might contribute into beneficial agronomic traits such as stress resistance or yield. The content of varying genomic regions in the pan-genome could include protein-coding genes as well as microRNA(miRNAs), small non-coding RNAs playing key roles in the regulation of gene expression. In this study, we performed in silico miRNA identification from the genomic sequences of 54 lineages of Brachypodium distachyon, aiming to explore varying miRNA contents and their functional interactions. A total of 115 miRNA families were identified in 54 lineages, 56 of which were found to be present in all lineages. The miRNA families were classified based on their conservation among lineages and potential mRNA targets were identified. Obtaining information about regulatory mechanisms stemming from these miRNAs offers strong potential to provide a better insight into the complex traits that were potentially present in some lineages. Future work could lead us to introduce these traits to different lineages or other economically important plant species in order to promote their survival in different environmental conditions.
Keywords: Brachypodium; microRNA; pan-genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Exploring the interaction between small RNAs and R genes during Brachypodium response to Fusarium culmorum infection.Gene. 2014 Feb 25;536(2):254-64. doi: 10.1016/j.gene.2013.12.025. Epub 2013 Dec 22. Gene. 2014. PMID: 24368332
-
In silico screening of the chicken genome for overlaps between genomic regions: microRNA genes, coding and non-coding transcriptional units, QTL, and genetic variations.Chromosome Res. 2016 May;24(2):225-30. doi: 10.1007/s10577-016-9517-9. Epub 2016 Jan 22. Chromosome Res. 2016. PMID: 26800695
-
Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in Brachypodium distachyon.BMC Genomics. 2016 Aug 15;17(1):636. doi: 10.1186/s12864-016-2968-8. BMC Genomics. 2016. PMID: 27527343 Free PMC article.
-
Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure.Nat Commun. 2017 Dec 19;8(1):2184. doi: 10.1038/s41467-017-02292-8. Nat Commun. 2017. PMID: 29259172 Free PMC article.
-
Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L.BMC Plant Biol. 2018 Dec 6;18(1):336. doi: 10.1186/s12870-018-1559-z. BMC Plant Biol. 2018. PMID: 30522432 Free PMC article.
Cited by
-
Combined Analysis of MicroRNAs and Target Genes Revealed miR156-SPLs and miR172-AP2 Are Involved in a Delayed Flowering Phenomenon After Chromosome Doubling in Black Goji (Lycium ruthencium).Front Genet. 2021 Jul 15;12:706930. doi: 10.3389/fgene.2021.706930. eCollection 2021. Front Genet. 2021. PMID: 34335704 Free PMC article.
-
Noncoding elements in wheat defence response to fusarium head blight.Sci Rep. 2025 Apr 30;15(1):15167. doi: 10.1038/s41598-025-00067-6. Sci Rep. 2025. PMID: 40307260 Free PMC article.
-
Comparative Analysis of Coding and Non-Coding Features within Insect Tolerance Loci in Wheat with Their Homologs in Cereal Genomes.Int J Mol Sci. 2021 Nov 16;22(22):12349. doi: 10.3390/ijms222212349. Int J Mol Sci. 2021. PMID: 34830231 Free PMC article.
References
LinkOut - more resources
Full Text Sources

