The Transcriptome of SH-SY5Y at Single-Cell Resolution: A CITE-Seq Data Analysis Workflow
- PMID: 34066513
- PMCID: PMC8163004
- DOI: 10.3390/mps4020028
The Transcriptome of SH-SY5Y at Single-Cell Resolution: A CITE-Seq Data Analysis Workflow
Abstract
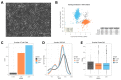
Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) is a recently established multimodal single cell analysis technique combining the immunophenotyping capabilities of antibody labeling and cell sorting with the resolution of single-cell RNA sequencing (scRNA-seq). By simply adding a 12-bp nucleotide barcode to antibodies (cell hashing), CITE-seq can be used to sequence antibody-bound tags alongside the cellular mRNA, thus reducing costs of scRNA-seq by performing it at the same time on multiple barcoded samples in a single run. Here, we illustrate an ideal CITE-seq data analysis workflow by characterizing the transcriptome of SH-SY5Y neuroblastoma cell line, a widely used model to study neuronal function and differentiation. We obtained transcriptomes from a total of 2879 single cells, measuring an average of 1600 genes/cell. Along with standard scRNA-seq data handling procedures, such as quality checks and cell filtering procedures, we performed exploratory analyses to identify most stable genes to be possibly used as reference housekeeping genes in qPCR experiments. We also illustrate how to use some popular R packages to investigate cell heterogeneity in scRNA-seq data, namely Seurat, Monocle, and slalom. Both the CITE-seq dataset and the code used to analyze it are freely shared and fully reusable for future research.
Keywords: CITE-seq; gene regulatory networks; neuroblastoma; single-cell; transcriptomics; unsupervised learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Angerer P., Simon L., Tritschler S., Wolf F.A., Fischer D., Theis F.J. Single Cells Make Big Data: New Challenges and Opportunities in Transcriptomics. Curr. Opin. Syst. Biol. 2017;4:85–91. doi: 10.1016/j.coisb.2017.07.004. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

