Small Molecule HIV-1 Attachment Inhibitors: Discovery, Mode of Action and Structural Basis of Inhibition
- PMID: 34066522
- PMCID: PMC8148533
- DOI: 10.3390/v13050843
Small Molecule HIV-1 Attachment Inhibitors: Discovery, Mode of Action and Structural Basis of Inhibition
Abstract
Viral entry into host cells is a critical step in the viral life cycle. HIV-1 entry is mediated by the sole surface envelope glycoprotein Env and is initiated by the interaction between Env and the host receptor CD4. This interaction, referred to as the attachment step, has long been considered an attractive target for inhibitor discovery and development. Fostemsavir, recently approved by the FDA, represents the first-in-class drug in the attachment inhibitor class. This review focuses on the discovery of temsavir (the active compound of fostemsavir) and analogs, mechanistic studies that elucidated the mode of action, and structural studies that revealed atomic details of the interaction between HIV-1 Env and attachment inhibitors. Challenges associated with emerging resistance mutations to the attachment inhibitors and the development of next-generation attachment inhibitors are also highlighted.
Keywords: HIV-1 entry; antiviral activity; attachment inhibitors; crystal structures; fostemsavir; temsavir.
Conflict of interest statement
The author declares no conflict of interest.
Figures
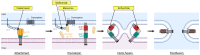


Similar articles
-
Viral Drug Resistance Through 48 Weeks, in a Phase 2b, Randomized, Controlled Trial of the HIV-1 Attachment Inhibitor Prodrug, Fostemsavir.J Acquir Immune Defic Syndr. 2018 Mar 1;77(3):299-307. doi: 10.1097/QAI.0000000000001602. J Acquir Immune Defic Syndr. 2018. PMID: 29206721 Free PMC article. Clinical Trial.
-
Fostemsavir: First Approval.Drugs. 2020 Sep;80(14):1485-1490. doi: 10.1007/s40265-020-01386-w. Drugs. 2020. PMID: 32852743 Review.
-
Neutralization Synergy between HIV-1 Attachment Inhibitor Fostemsavir and Anti-CD4 Binding Site Broadly Neutralizing Antibodies against HIV.J Virol. 2019 Feb 5;93(4):e01446-18. doi: 10.1128/JVI.01446-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30518644 Free PMC article.
-
Composition and Orientation of the Core Region of Novel HIV-1 Entry Inhibitors Influences Metabolic Stability.Molecules. 2020 Mar 21;25(6):1430. doi: 10.3390/molecules25061430. Molecules. 2020. PMID: 32245167 Free PMC article.
-
Ibalizumab and Fostemsavir in the Management of Heavily Pre-Treated HIV-infected Patients.Recent Pat Antiinfect Drug Discov. 2018;13(3):190-197. doi: 10.2174/1574891X13666181031120019. Recent Pat Antiinfect Drug Discov. 2018. PMID: 30378502 Review.
Cited by
-
In Vitro Anti-HIV-1 Activity of Fucoidans from Brown Algae.Mar Drugs. 2024 Jul 31;22(8):355. doi: 10.3390/md22080355. Mar Drugs. 2024. PMID: 39195471 Free PMC article.
-
The Role of Peptides in Combatting HIV Infection: Applications and Insights.Molecules. 2024 Oct 19;29(20):4951. doi: 10.3390/molecules29204951. Molecules. 2024. PMID: 39459319 Free PMC article. Review.
-
Identification of human immunodeficiency virus -1 E protein-targeting lead compounds by pharmacophore based screening.Saudi Med J. 2022 Dec;43(12):1324-1332. doi: 10.15537/smj.2022.43.12.20220599. Saudi Med J. 2022. PMID: 36517066 Free PMC article.
-
Synthetic Approaches to Piperazine-Containing Drugs Approved by FDA in the Period of 2011-2023.Molecules. 2023 Dec 21;29(1):68. doi: 10.3390/molecules29010068. Molecules. 2023. PMID: 38202651 Free PMC article. Review.
-
Peptide Triazole Inhibitors of HIV-1: Hijackers of Env Metastability.Curr Protein Pept Sci. 2023;24(1):59-77. doi: 10.2174/1389203723666220610120927. Curr Protein Pept Sci. 2023. PMID: 35692162 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

