The Role of Structural Variation in Adaptation and Evolution of Yeast and Other Fungi
- PMID: 34066718
- PMCID: PMC8150848
- DOI: 10.3390/genes12050699
The Role of Structural Variation in Adaptation and Evolution of Yeast and Other Fungi
Abstract
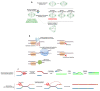
Mutations in DNA can be limited to one or a few nucleotides, or encompass larger deletions, insertions, duplications, inversions and translocations that span long stretches of DNA or even full chromosomes. These so-called structural variations (SVs) can alter the gene copy number, modify open reading frames, change regulatory sequences or chromatin structure and thus result in major phenotypic changes. As some of the best-known examples of SV are linked to severe genetic disorders, this type of mutation has traditionally been regarded as negative and of little importance for adaptive evolution. However, the advent of genomic technologies uncovered the ubiquity of SVs even in healthy organisms. Moreover, experimental evolution studies suggest that SV is an important driver of evolution and adaptation to new environments. Here, we provide an overview of the causes and consequences of SV and their role in adaptation, with specific emphasis on fungi since these have proven to be excellent models to study SV.
Keywords: adaptation; fungi; structural variation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Geographic distribution and adaptive significance of genomic structural variants: an anthropological genetics perspective.Hum Biol. 2014 Fall;86(4):260-75. doi: 10.13110/humanbiology.86.4.0260. Hum Biol. 2014. PMID: 25959693 Review.
-
Exploring the role of polymorphic interspecies structural variants in reproductive isolation and adaptive divergence in Eucalyptus.Gigascience. 2024 Jan 2;13:giae029. doi: 10.1093/gigascience/giae029. Gigascience. 2024. PMID: 38869149 Free PMC article.
-
Whole-genome sequencing with long reads reveals complex structure and origin of structural variation in human genetic variations and somatic mutations in cancer.Genome Med. 2021 Apr 29;13(1):65. doi: 10.1186/s13073-021-00883-1. Genome Med. 2021. PMID: 33910608 Free PMC article.
-
Genomic structural variation: A complex but important driver of human evolution.Am J Biol Anthropol. 2023 Aug;181 Suppl 76(Suppl 76):118-144. doi: 10.1002/ajpa.24713. Epub 2023 Feb 16. Am J Biol Anthropol. 2023. PMID: 36794631 Free PMC article. Review.
-
The landscape and predicted roles of structural variants in Fusarium graminearum genomes.G3 (Bethesda). 2024 Jun 5;14(6):jkae065. doi: 10.1093/g3journal/jkae065. G3 (Bethesda). 2024. PMID: 38546739 Free PMC article.
Cited by
-
Genomic Adaptations of Saccharomyces Genus to Wine Niche.Microorganisms. 2022 Sep 9;10(9):1811. doi: 10.3390/microorganisms10091811. Microorganisms. 2022. PMID: 36144411 Free PMC article. Review.
-
Compensatory Genetic and Transcriptional Cytonuclear Coordination in Allopolyploid Lager Yeast (Saccharomyces pastorianus).Mol Biol Evol. 2022 Nov 3;39(11):msac228. doi: 10.1093/molbev/msac228. Mol Biol Evol. 2022. PMID: 36260528 Free PMC article.
-
Recent insights into the evolution of mutation rates in yeast.Curr Opin Genet Dev. 2022 Oct;76:101953. doi: 10.1016/j.gde.2022.101953. Epub 2022 Jul 11. Curr Opin Genet Dev. 2022. PMID: 35834945 Free PMC article. Review.
-
Domestication signatures in the non-conventional yeast Lachancea cidri.mSystems. 2024 Jan 23;9(1):e0105823. doi: 10.1128/msystems.01058-23. Epub 2023 Dec 12. mSystems. 2024. PMID: 38085042 Free PMC article.
-
Identifying Targets of Selection in Laboratory Evolution Experiments.J Mol Evol. 2023 Jun;91(3):345-355. doi: 10.1007/s00239-023-10096-2. Epub 2023 Feb 21. J Mol Evol. 2023. PMID: 36810618 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

