Identification of Bile Salt Hydrolase and Bile Salt Resistance in a Probiotic Bacterium Lactobacillus gasseri JCM1131T
- PMID: 34066735
- PMCID: PMC8151060
- DOI: 10.3390/microorganisms9051011
Identification of Bile Salt Hydrolase and Bile Salt Resistance in a Probiotic Bacterium Lactobacillus gasseri JCM1131T
Abstract
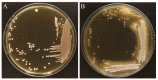
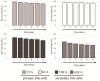
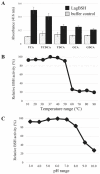
Lactobacillus gasseri is one of the most likely probiotic candidates among many Lactobacillus species. Although bile salt resistance has been defined as an important criterion for selection of probiotic candidates since it allows probiotic bacteria to survive in the gut, both its capability and its related enzyme, bile salt hydrolase (BSH), in L. gasseri is still largely unknown. Here, we report that the well-known probiotic bacterium L. gasseri JCM1131T possesses BSH activity and bile salt resistance capability. Indeed, this strain apparently showed BSH activity on the plate assay and highly tolerated the primary bile salts and even taurine-conjugated secondary bile salt. We further isolated a putative BSH enzyme (LagBSH) from strain JCM1131T and characterized the enzymatic function. The purified LagBSH protein exhibited quite high deconjugation activity for taurocholic acid and taurochenodeoxycholic acid. The lagBSH gene was constitutively expressed in strain JCM1131T, suggesting that LagBSH likely contributes to bile salt resistance of the strain and may be associated with survival capability of strain JCM1131T within the human intestine by bile detoxification. Thus, this study first demonstrated the bile salt resistance and its responsible enzyme (BSH) activity in strain JCM1131T, which further supports the importance of the typical lactic acid bacterium as probiotics.
Keywords: Lactobacillus gasseri; Ntn-hydrolase family protein; bile salt hydrolase; probiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

