Hepatitis B Virus Pre-S Gene Deletions and Pre-S Deleted Proteins: Clinical and Molecular Implications in Hepatocellular Carcinoma
- PMID: 34066744
- PMCID: PMC8151789
- DOI: 10.3390/v13050862
Hepatitis B Virus Pre-S Gene Deletions and Pre-S Deleted Proteins: Clinical and Molecular Implications in Hepatocellular Carcinoma
Abstract
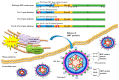
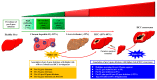
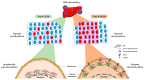
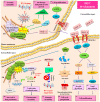
Hepatocellular carcinoma (HCC) is one of the most frequent and fatal human cancers worldwide and its development and prognosis are intimately associated with chronic infection with hepatitis B virus (HBV). The identification of genetic mutations and molecular mechanisms that mediate HBV-induced tumorigenesis therefore holds promise for the development of potential biomarkers and targets for HCC prevention and therapy. The presence of HBV pre-S gene deletions in the blood and the expression of pre-S deleted proteins in the liver tissues of patients with chronic hepatitis B and HBV-related HCC have emerged as valuable biomarkers for higher incidence rates of HCC development and a higher risk of HCC recurrence after curative surgical resection, respectively. Moreover, pre-S deleted proteins are regarded as important oncoproteins that activate multiple signaling pathways to induce DNA damage and promote growth and proliferation in hepatocytes, leading to HCC development. The signaling molecules dysregulated by pre-S deleted proteins have also been validated as potential targets for the prevention of HCC development. In this review, we summarize the clinical and molecular implications of HBV pre-S gene deletions and pre-S deleted proteins in HCC development and recurrence and highlight their potential applications in HCC prevention and therapy.
Keywords: biomarkers; hepatitis B virus; hepatocellular carcinoma; pre-S deleted proteins; pre-S gene deletions; targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

