OBP14 (Odorant-Binding Protein) Sensing in Adelphocoris lineolatus Based on Peptide Nucleic Acid and Graphene Oxide
- PMID: 34066819
- PMCID: PMC8151863
- DOI: 10.3390/insects12050422
OBP14 (Odorant-Binding Protein) Sensing in Adelphocoris lineolatus Based on Peptide Nucleic Acid and Graphene Oxide
Abstract
OBPs play a crucial role in the recognition of ligands and are involved in the initial steps of semiochemical perception. The diverse expression of OBP genes allows them to participate in different physiological functions in insects. In contrast to classic OBPs with typical olfactory roles in A. lineolatus, the physiological functions of Plus-C OBPs remain largely unknown. In addition, detection of the expression of insect OBP genes by conventional methods is difficult in vitro. Here, we focused on AlinOBP14, a Plus-C OBP from A. lineolatus, and we developed a PNA-GO-based mRNA biosensor to detect the expression of AlinOBP14. The results demonstrated that AlinOBP14 plays dual roles in A. lineolatus. The AlinOBP14 is expressed beneath the epidermis of the vertex and gena in heads of A. lineolatus, and it functions as a carrier for three terpenoids, while AlinOBP14 is also expressed in the peripheral antennal lobe and functions as a carrier for endogenous compounds such as precursors for juvenile hormone (JH) and JHⅢ. Our investigation provides a new method to detect the expression of OBP genes in insects, and the technique will facilitate the use of these genes as potential targets for novel insect behavioral regulation strategies against the pest.
Keywords: Adelphocoris lineolatus; AlinOBP14; Plus-C OBPs; graphene oxide (GO); in vitro; in vivo; juvenile hormone; mRNA biosensor; peptide nucleic acid (PNA); terpenoids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
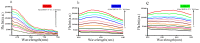
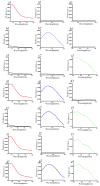
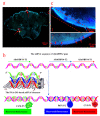
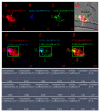

Similar articles
-
Identification and Characterization of Odorant Binding Proteins in the Forelegs of Adelphocoris lineolatus (Goeze).Front Physiol. 2017 Sep 26;8:735. doi: 10.3389/fphys.2017.00735. eCollection 2017. Front Physiol. 2017. PMID: 29018358 Free PMC article.
-
Expression patterns and ligand binding characterization of Plus-C odorant-binding protein 14 from Adelphocoris lineolatus (Goeze).Comp Biochem Physiol B Biochem Mol Biol. 2019 Jan;227:75-82. doi: 10.1016/j.cbpb.2018.10.001. Epub 2018 Oct 5. Comp Biochem Physiol B Biochem Mol Biol. 2019. PMID: 30292754
-
Identification and tissue distribution of odorant binding protein genes in the lucerne plant bug Adelphocoris lineolatus (Goeze).Insect Biochem Mol Biol. 2011 Apr;41(4):254-63. doi: 10.1016/j.ibmb.2011.01.002. Epub 2011 Jan 11. Insect Biochem Mol Biol. 2011. PMID: 21232599
-
An overview of odorant-binding protein functions in insect peripheral olfactory reception.Genet Mol Res. 2011 Dec 8;10(4):3056-69. doi: 10.4238/2011.December.8.2. Genet Mol Res. 2011. PMID: 22180039 Review.
-
Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis.Front Physiol. 2018 Aug 24;9:1163. doi: 10.3389/fphys.2018.01163. eCollection 2018. Front Physiol. 2018. PMID: 30197600 Free PMC article. Review.
Cited by
-
Function and Characterization Analysis of BodoOBP8 from Bradysia odoriphaga (Diptera: Sciaridae) in the Recognition of Plant Volatiles and Sex Pheromones.Insects. 2021 Sep 28;12(10):879. doi: 10.3390/insects12100879. Insects. 2021. PMID: 34680648 Free PMC article.
-
Label-Free DNA Biosensor Based on Reduced Graphene Oxide and Gold Nanoparticles.Biosensors (Basel). 2023 Aug 8;13(8):797. doi: 10.3390/bios13080797. Biosensors (Basel). 2023. PMID: 37622883 Free PMC article.
-
Identification of Odorant-Binding and Chemosensory Protein Genes in Mythimna separata Adult Brains Using Transcriptome Analyses.Front Physiol. 2022 Feb 28;13:839559. doi: 10.3389/fphys.2022.839559. eCollection 2022. Front Physiol. 2022. PMID: 35295575 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

