Peripheral Membrane Proteins: Promising Therapeutic Targets across Domains of Life
- PMID: 34066904
- PMCID: PMC8151925
- DOI: 10.3390/membranes11050346
Peripheral Membrane Proteins: Promising Therapeutic Targets across Domains of Life
Abstract
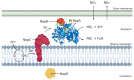
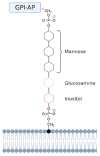
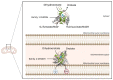
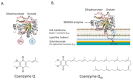
Membrane proteins can be classified into two main categories-integral and peripheral membrane proteins-depending on the nature of their membrane interaction. Peripheral membrane proteins are highly unique amphipathic proteins that interact with the membrane indirectly, using electrostatic or hydrophobic interactions, or directly, using hydrophobic tails or GPI-anchors. The nature of this interaction not only influences the location of the protein in the cell, but also the function. In addition to their unique relationship with the cell membrane, peripheral membrane proteins often play a key role in the development of human diseases such as African sleeping sickness, cancer, and atherosclerosis. This review will discuss the membrane interaction and role of periplasmic nitrate reductase, CymA, cytochrome c, alkaline phosphatase, ecto-5'-nucleotidase, acetylcholinesterase, alternative oxidase, type-II NADH dehydrogenase, and dihydroorotate dehydrogenase in certain diseases. The study of these proteins will give new insights into their function and structure, and may ultimately lead to ground-breaking advances in the treatment of severe diseases.
Keywords: GPI-anchored proteins; drug targets; electrostatic interactions; human diseases; hydrophobic membrane anchor; peripheral membrane proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Glycosyl-phosphatidylinositol-anchored membrane proteins.J Am Soc Nephrol. 1992 Oct;3(4):895-906. doi: 10.1681/ASN.V34895. J Am Soc Nephrol. 1992. PMID: 1450366
-
Glycosyl-phosphatidylinositol anchored membrane enzymes.Clin Chim Acta. 1997 Oct 9;266(1):3-12. doi: 10.1016/s0009-8981(97)00161-7. Clin Chim Acta. 1997. PMID: 9435983
-
4'-Amino-benzamido-taurocholic acid selectively solubilizes glycosyl-phosphatidylinositol-anchored membrane proteins and improves lipolytic cleavage of their membrane anchors by specific phospholipases.Arch Biochem Biophys. 1994 Mar;309(2):329-40. doi: 10.1006/abbi.1994.1121. Arch Biochem Biophys. 1994. PMID: 8135545
-
The membrane-bound tetrahaem c-type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella.Biochem Soc Trans. 2002 Aug;30(4):658-62. doi: 10.1042/bst0300658. Biochem Soc Trans. 2002. PMID: 12196158 Review.
-
The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins.Biochemistry. 2008 Jul 8;47(27):6991-7000. doi: 10.1021/bi8006324. Epub 2008 Jun 17. Biochemistry. 2008. PMID: 18557633 Free PMC article. Review.
Cited by
-
DREAMM: a web-based server for drugging protein-membrane interfaces as a novel workflow for targeted drug design.Bioinformatics. 2022 Dec 13;38(24):5449-5451. doi: 10.1093/bioinformatics/btac680. Bioinformatics. 2022. PMID: 36355565 Free PMC article.
-
Whole Proteome-Based Therapeutic Targets Annotation and Designing of Multi-Epitope-Based Vaccines against the Gram-Negative XDR-Alcaligenes faecalis Bacterium.Vaccines (Basel). 2022 Mar 17;10(3):462. doi: 10.3390/vaccines10030462. Vaccines (Basel). 2022. PMID: 35335094 Free PMC article.
-
Membrane Proteins and Membrane Curvature: Mutual Interactions and a Perspective on Disease Treatments.Biomolecules. 2023 Dec 11;13(12):1772. doi: 10.3390/biom13121772. Biomolecules. 2023. PMID: 38136643 Free PMC article. Review.
-
Machine learning in computational modelling of membrane protein sequences and structures: From methodologies to applications.Comput Struct Biotechnol J. 2023 Jan 28;21:1205-1226. doi: 10.1016/j.csbj.2023.01.036. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36817959 Free PMC article. Review.
-
Infrared Photoactivation Enables Improved Native Top-Down Mass Spectrometry of Transmembrane Proteins.Anal Chem. 2023 Sep 5;95(35):13361-13367. doi: 10.1021/acs.analchem.3c02788. Epub 2023 Aug 23. Anal Chem. 2023. PMID: 37610409 Free PMC article.
References
-
- Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J. Molecular Cell Biology. W.H. Freeman; New York, NY, USA: 2000. Membrane Proteins.
-
- Protein Data Bank RCSB PDB-5T4Q: Autoinhibited E. coli ATP Synthase State 3. [(accessed on 1 April 2021)];2016 Available online: https://www.rcsb.org/structure/5t4q.
-
- Protein Data Bank RCSB PDB-2J7A: Crystal Structure of Cytochrome C Nitrite Reductase NrfHA Complex from Desulfovibrio vulgaris. [(accessed on 1 April 2021)];2006 Available online: https://www.rcsb.org/structure/2j7a.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

