Codon Bias Can Determine Sorting of a Potassium Channel Protein
- PMID: 34066987
- PMCID: PMC8151079
- DOI: 10.3390/cells10051128
Codon Bias Can Determine Sorting of a Potassium Channel Protein
Abstract
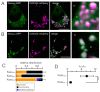
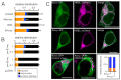
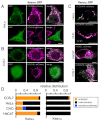
Due to the redundancy of the genetic code most amino acids are encoded by multiple synonymous codons. It has been proposed that a biased frequency of synonymous codons can affect the function of proteins by modulating distinct steps in transcription, translation and folding. Here, we use two similar prototype K+ channels as model systems to examine whether codon choice has an impact on protein sorting. By monitoring transient expression of GFP-tagged channels in mammalian cells, we find that one of the two channels is sorted in a codon and cell cycle-dependent manner either to mitochondria or the secretory pathway. The data establish that a gene with either rare or frequent codons serves, together with a cell-state-dependent decoding mechanism, as a secondary code for sorting intracellular membrane proteins.
Keywords: codon usage; dual sorting; effect of synonymous codon exchange; membrane protein sorting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

