Homologous Recombination Repair Deficiency and Implications for Tumor Immunogenicity
- PMID: 34067105
- PMCID: PMC8124836
- DOI: 10.3390/cancers13092249
Homologous Recombination Repair Deficiency and Implications for Tumor Immunogenicity
Abstract
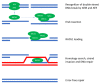
Homologous recombination repair deficiency (HRD) can be observed in virtually all cancer types. Although HRD sensitizes tumors to DNA-damaging chemotherapy and poly(ADP-ribose) polymerase (PARP) inhibitors, all patients ultimately develop resistance to these therapies. Therefore, it is necessary to identify therapeutic regimens with a more durable efficacy. HRD tumors have been suggested to be more immunogenic and, therefore, more susceptible to treatment with checkpoint inhibitors. In this review, we describe how HRD might mechanistically affect antitumor immunity and summarize the available translational evidence for an association between HRD and antitumor immunity across multiple tumor types. In addition, we give an overview of all available clinical data on the efficacy of checkpoint inhibitors in HRD tumors and describe the evidence for using treatment strategies that combine checkpoint inhibitors with PARP inhibitors.
Keywords: cancer; homologous recombination repair deficiency; immune checkpoint inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

