Mechanisms That Activate 26S Proteasomes and Enhance Protein Degradation
- PMID: 34067263
- PMCID: PMC8224753
- DOI: 10.3390/biom11060779
Mechanisms That Activate 26S Proteasomes and Enhance Protein Degradation
Abstract
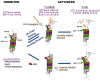
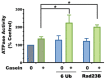
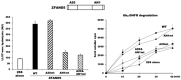
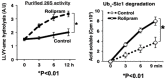
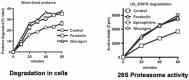
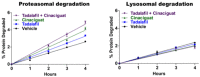
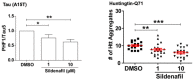
Although ubiquitination is widely assumed to be the only regulated step in the ubiquitin-proteasome pathway, recent studies have demonstrated several important mechanisms that regulate the activities of the 26S proteasome. Most proteasomes in cells are inactive but, upon binding a ubiquitinated substrate, become activated by a two-step mechanism requiring an association of the ubiquitin chain with Usp14 and then a loosely folded protein domain with the ATPases. The initial activation step is signaled by Usp14's UBL domain, and many UBL-domain-containing proteins (e.g., Rad23, Parkin) also activate the proteasome. ZFAND5 is a distinct type of activator that binds ubiquitin conjugates and the proteasome and stimulates proteolysis during muscle atrophy. The proteasome's activities are also regulated through subunit phosphorylation. Agents that raise cAMP and activate PKA stimulate within minutes Rpn6 phosphorylation and enhance the selective degradation of short-lived proteins. Likewise, hormones, fasting, and exercise, which raise cAMP, activate proteasomes and proteolysis in target tissues. Agents that raise cGMP and activate PKG also stimulate 26S activities but modify different subunit(s) and stimulate also the degradation of long-lived cell proteins. Both kinases enhance the selective degradation of aggregation-prone proteins that cause neurodegenerative diseases. These new mechanisms regulating proteolysis thus have clear physiological importance and therapeutic potential.
Keywords: PKA; PKG; Rad23; UBL-domain-containing proteins; Usp14; ZFAND5; ubiquitin–proteasome system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

