Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images
- PMID: 34067726
- PMCID: PMC8156071
- DOI: 10.3390/cancers13102419
Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images
Abstract
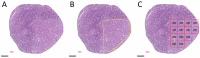
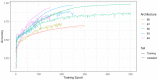
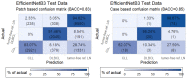
The diagnosis and the subtyping of non-Hodgkin lymphoma (NHL) are challenging and require expert knowledge, great experience, thorough morphological analysis, and often additional expensive immunohistological and molecular methods. As these requirements are not always available, supplemental methods supporting morphological-based decision making and potentially entity subtyping are required. Deep learning methods have been shown to classify histopathological images with high accuracy, but data on NHL subtyping are limited. After annotation of histopathological whole-slide images and image patch extraction, we trained and optimized an EfficientNet convolutional neuronal network algorithm on 84,139 image patches from 629 patients and evaluated its potential to classify tumor-free reference lymph nodes, nodal small lymphocytic lymphoma/chronic lymphocytic leukemia, and nodal diffuse large B-cell lymphoma. The optimized algorithm achieved an accuracy of 95.56% on an independent test set including 16,960 image patches from 125 patients after the application of quality controls. Automatic classification of NHL is possible with high accuracy using deep learning on histopathological images and routine diagnostic applications should be pursued.
Keywords: CLL/SLL; CNN; DLBCL; artificial intelligence; deep learning; histopathology.
Conflict of interest statement
No external funding has been received. The authors declare no conflict of interest.
Figures


Similar articles
-
Deep Learning for the Classification of Small-Cell and Non-Small-Cell Lung Cancer.Cancers (Basel). 2020 Jun 17;12(6):1604. doi: 10.3390/cancers12061604. Cancers (Basel). 2020. PMID: 32560475 Free PMC article.
-
Classification of digital pathological images of non-Hodgkin's lymphoma subtypes based on the fusion of transfer learning and principal component analysis.Med Phys. 2020 Sep;47(9):4241-4253. doi: 10.1002/mp.14357. Epub 2020 Jul 18. Med Phys. 2020. PMID: 32593219
-
Automated Diagnosis of Lymphoma with Digital Pathology Images Using Deep Learning.Ann Clin Lab Sci. 2019 Mar;49(2):153-160. Ann Clin Lab Sci. 2019. PMID: 31028058
-
Learning from the failures of drug discovery in B-cell non-Hodgkin lymphomas and perspectives for the future: chronic lymphocytic leukemia and diffuse large B-cell lymphoma as two ends of a spectrum in drug development.Expert Opin Drug Discov. 2017 Jul;12(7):733-745. doi: 10.1080/17460441.2017.1329293. Epub 2017 Jun 7. Expert Opin Drug Discov. 2017. PMID: 28494631 Review.
-
[Non-Hodgkin's lymphoma. Cytology and cytochemistry].Veroff Pathol. 1986;123:1-241. Veroff Pathol. 1986. PMID: 3518274 Review. German.
Cited by
-
Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images.Cancers (Basel). 2023 Oct 29;15(21):5205. doi: 10.3390/cancers15215205. Cancers (Basel). 2023. PMID: 37958379 Free PMC article.
-
Artificial intelligence performance in detecting lymphoma from medical imaging: a systematic review and meta-analysis.BMC Med Inform Decis Mak. 2024 Jan 8;24(1):13. doi: 10.1186/s12911-023-02397-9. BMC Med Inform Decis Mak. 2024. PMID: 38191361 Free PMC article.
-
Morphology and multiparameter flow cytometry combined for integrated lymphoma diagnosis on small volume samples: possibilities and limitations.Virchows Arch. 2024 Oct;485(4):591-604. doi: 10.1007/s00428-024-03819-3. Epub 2024 May 28. Virchows Arch. 2024. PMID: 38805049 Free PMC article. Review.
-
Role of artificial intelligence in haematolymphoid diagnostics.Histopathology. 2025 Jan;86(1):58-68. doi: 10.1111/his.15327. Epub 2024 Oct 22. Histopathology. 2025. PMID: 39435690 Free PMC article. Review.
-
Application of EfficientNet-B0 and GRU-based deep learning on classifying the colposcopy diagnosis of precancerous cervical lesions.Cancer Med. 2023 Apr;12(7):8690-8699. doi: 10.1002/cam4.5581. Epub 2023 Jan 11. Cancer Med. 2023. PMID: 36629131 Free PMC article.
References
-
- National Cancer Institute Cancer Stat Facts: Non-Hodgkin Lymphoma. [(accessed on 21 January 2021)]; Available online: https://seer.cancer.gov/statfacts/html/nhl.html.
-
- Swerdlow S.H., Campo E., Pileri S.A., Harris N.L., Stein H., Siebert R., Advani R., Ghielmini M., Salles G.A., Zelenetz A.D., et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. - DOI - PMC - PubMed
-
- Di Napoli A., Remotti D., Agostinelli C., Ambrosio M.R., Ascani S., Carbone A., Facchetti F., Lazzi S., Leoncini L., Lucioni M., et al. A practical algorithmic approach to mature aggressive B cell lymphoma diagnosis in the double/triple hit era: Selecting cases, matching clinical benefit. Virchows Archiv. 2019;475:513–518. doi: 10.1007/s00428-019-02637-2. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

