Understanding LAG-3 Signaling
- PMID: 34067904
- PMCID: PMC8156499
- DOI: 10.3390/ijms22105282
Understanding LAG-3 Signaling
Abstract
Lymphocyte activation gene 3 (LAG-3) is a cell surface inhibitory receptor with multiple biological activities over T cell activation and effector functions. LAG-3 plays a regulatory role in immunity and emerged some time ago as an inhibitory immune checkpoint molecule comparable to PD-1 and CTLA-4 and a potential target for enhancing anti-cancer immune responses. LAG-3 is the third inhibitory receptor to be exploited in human anti-cancer immunotherapies, and it is considered a potential next-generation cancer immunotherapy target in human therapy, right next to PD-1 and CTLA-4. Unlike PD-1 and CTLA-4, the exact mechanisms of action of LAG-3 and its relationship with other immune checkpoint molecules remain poorly understood. This is partly caused by the presence of non-conventional signaling motifs in its intracellular domain that are different from other conventional immunoregulatory signaling motifs but with similar inhibitory activities. Here we summarize the current understanding of LAG-3 signaling and its role in LAG-3 functions, from its mechanisms of action to clinical applications.
Keywords: LAG-3; cancer signaling; immune checkpoint; immunotherapy; targeted therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
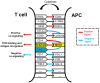
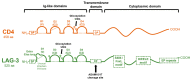


References
Publication types
MeSH terms
Substances
Grants and funding
- PROYE16001ESCO/Fundación Científica Asociación Española Contra el Cáncer
- FIS PI17/02119/Instituto de Salud Carlos III
- FIS PI20/00010/Instituto de Salud Carlos III
- BMED 050-2019/Departamento de Salud, Gobierno de Navarra
- AGATA, Ref 0011-1411-2020-000013/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra
- LINTERNA, Ref. 0011-1411-2020-000033/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra
- DESCARTHES, 0011-1411-2019-000058/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra
- ISOLDA; ID: 848166/Horizon 2020 Framework Programme
- TRANSPOCART ICI19/00069/Instituto de Salud Carlos III
- COV20/00000/Instituto de Salud Carlos III
LinkOut - more resources
Full Text Sources
Other Literature Sources

