Atovaquone Suppresses Triple-Negative Breast Tumor Growth by Reducing Immune-Suppressive Cells
- PMID: 34068008
- PMCID: PMC8152242
- DOI: 10.3390/ijms22105150
Atovaquone Suppresses Triple-Negative Breast Tumor Growth by Reducing Immune-Suppressive Cells
Abstract
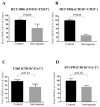
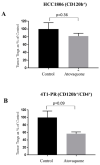
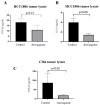
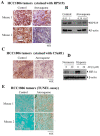
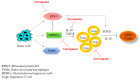
A major contributing factor in triple-negative breast cancer progression is its ability to evade immune surveillance. One mechanism for this immunosuppression is through ribosomal protein S19 (RPS19), which facilitates myeloid-derived suppressor cells (MDSCs) recruitment in tumors, which generate cytokines TGF-β and IL-10 and induce regulatory T cells (Tregs), all of which are immunosuppressive and enhance tumor progression. Hence, enhancing the immune system in breast tumors could be a strategy for anticancer therapeutics. The present study evaluated the immune response of atovaquone, an antiprotozoal drug, in three independent breast-tumor models. Our results demonstrated that oral administration of atovaquone reduced HCC1806, CI66 and 4T1 paclitaxel-resistant (4T1-PR) breast-tumor growth by 45%, 70% and 42%, respectively. MDSCs, TGF-β, IL-10 and Tregs of blood and tumors were analyzed from all of these in vivo models. Our results demonstrated that atovaquone treatment in mice bearing HCC1806 tumors reduced MDSCs from tumor and blood by 70% and 30%, respectively. We also observed a 25% reduction in tumor MDSCs in atovaquone-treated mice bearing CI66 and 4T1-PR tumors. In addition, a decrease in TGF-β and IL-10 in tumor lysates was observed in atovaquone-treated mice with a reduction in tumor Tregs. Moreover, a significant reduction in the expression of RPS19 was found in tumors treated with atovaquone.
Keywords: C5aR1; HIF1α; RPS19; atovaquone; cytokines; myeloid-derived tumor-suppressor cells; regulatory T cells; repurposing; triple-negative breast cancer.
Conflict of interest statement
The authors declare that there are no competing interest.
Figures

Similar articles
-
Therapeutic targeting with DABIL-4 depletes myeloid suppressor cells in 4T1 triple-negative breast cancer model.Mol Oncol. 2021 May;15(5):1330-1344. doi: 10.1002/1878-0261.12938. Epub 2021 Mar 24. Mol Oncol. 2021. PMID: 33682324 Free PMC article.
-
Atovaquone: An Antiprotozoal Drug Suppresses Primary and Resistant Breast Tumor Growth by Inhibiting HER2/β-Catenin Signaling.Mol Cancer Ther. 2019 Oct;18(10):1708-1720. doi: 10.1158/1535-7163.MCT-18-1286. Epub 2019 Jul 3. Mol Cancer Ther. 2019. PMID: 31270151 Free PMC article.
-
TGF-β Enhances the Anti-inflammatory Effect of Tumor- Infiltrating CD33+11b+HLA-DR Myeloid-Derived Suppressor Cells in Gastric Cancer: A Possible Relation to MicroRNA-494.Asian Pac J Cancer Prev. 2020 Nov 1;21(11):3393-3403. doi: 10.31557/APJCP.2020.21.11.3393. Asian Pac J Cancer Prev. 2020. PMID: 33247701 Free PMC article.
-
Suppression of T cells by myeloid-derived suppressor cells in cancer.Hum Immunol. 2017 Feb;78(2):113-119. doi: 10.1016/j.humimm.2016.12.001. Epub 2016 Dec 7. Hum Immunol. 2017. PMID: 27939507 Review.
-
Suppressive role of myeloid-derived suppressor cells (MDSCs) in the microenvironment of breast cancer and targeted immunotherapies.Oncotarget. 2016 Sep 27;7(39):64505-64511. doi: 10.18632/oncotarget.11352. Oncotarget. 2016. PMID: 27542274 Free PMC article. Review.
Cited by
-
Repurposing approved non-oncology drugs for cancer therapy: a comprehensive review of mechanisms, efficacy, and clinical prospects.Eur J Med Res. 2023 Sep 14;28(1):345. doi: 10.1186/s40001-023-01275-4. Eur J Med Res. 2023. PMID: 37710280 Free PMC article. Review.
-
Potentials of ribosomopathy gene as pharmaceutical targets for cancer treatment.J Pharm Anal. 2024 Mar;14(3):308-320. doi: 10.1016/j.jpha.2023.10.001. Epub 2023 Oct 13. J Pharm Anal. 2024. PMID: 38618250 Free PMC article. Review.
-
Novel therapeutic strategies targeting myeloid-derived suppressor cell immunosuppressive mechanisms for cancer treatment.Explor Target Antitumor Ther. 2024;5(1):187-207. doi: 10.37349/etat.2024.00212. Epub 2024 Feb 28. Explor Target Antitumor Ther. 2024. PMID: 38464388 Free PMC article. Review.
-
The Immune Landscape of Breast Cancer: Strategies for Overcoming Immunotherapy Resistance.Cancers (Basel). 2021 Nov 29;13(23):6012. doi: 10.3390/cancers13236012. Cancers (Basel). 2021. PMID: 34885122 Free PMC article. Review.
-
Reprogramming tumor immune microenvironment by milbemycin oxime results in pancreatic tumor growth suppression and enhanced anti-PD-1 efficacy.Mol Ther. 2024 Sep 4;32(9):3145-3162. doi: 10.1016/j.ymthe.2024.07.029. Epub 2024 Aug 3. Mol Ther. 2024. PMID: 39097773 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

