Principles and Practical Considerations for the Analysis of Disease-Associated Alternative Splicing Events Using the Gateway Cloning-Based Minigene Vectors pDESTsplice and pSpliceExpress
- PMID: 34068052
- PMCID: PMC8152502
- DOI: 10.3390/ijms22105154
Principles and Practical Considerations for the Analysis of Disease-Associated Alternative Splicing Events Using the Gateway Cloning-Based Minigene Vectors pDESTsplice and pSpliceExpress
Abstract
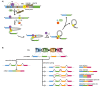
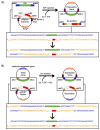
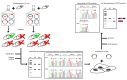
Splicing is an important RNA processing step. Genetic variations can alter the splicing process and thereby contribute to the development of various diseases. Alterations of the splicing pattern can be examined by gene expression analyses, by computational tools for predicting the effects of genetic variants on splicing, and by splicing reporter minigene assays for studying alternative splicing events under defined conditions. The minigene assay is based on transient transfection of cells with a vector containing a genomic region of interest cloned between two constitutive exons. Cloning can be accomplished by the use of restriction enzymes or by site-specific recombination using Gateway cloning. The vectors pDESTsplice and pSpliceExpress represent two minigene systems based on Gateway cloning, which are available through the Addgene plasmid repository. In this review, we describe the features of these two splicing reporter minigene systems. Moreover, we provide an overview of studies in which determinants of alternative splicing were investigated by using pDESTsplice or pSpliceExpress. The studies were reviewed with regard to the investigated splicing regulatory events and the experimental strategy to construct and perform a splicing reporter minigene assay. We further elaborate on how analyses on the regulation of RNA splicing offer promising prospects for gaining important insights into disease mechanisms.
Keywords: RNA processing; alternative splicing; gateway cloning; pDESTsplice; pSpliceExpress; splicing regulation; splicing reporter minigene assay.
Conflict of interest statement
The authors declare no conflict of interest in relation to this article.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

