Novel Roles of SH2 and SH3 Domains in Lipid Binding
- PMID: 34068055
- PMCID: PMC8152464
- DOI: 10.3390/cells10051191
Novel Roles of SH2 and SH3 Domains in Lipid Binding
Abstract
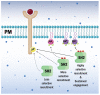
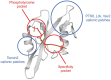
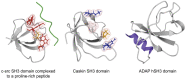
Signal transduction, the ability of cells to perceive information from the surroundings and alter behavior in response, is an essential property of life. Studies on tyrosine kinase action fundamentally changed our concept of cellular regulation. The induced assembly of subcellular hubs via the recognition of local protein or lipid modifications by modular protein interactions is now a central paradigm in signaling. Such molecular interactions are mediated by specific protein interaction domains. The first such domain identified was the SH2 domain, which was postulated to be a reader capable of finding and binding protein partners displaying phosphorylated tyrosine side chains. The SH3 domain was found to be involved in the formation of stable protein sub-complexes by constitutively attaching to proline-rich surfaces on its binding partners. The SH2 and SH3 domains have thus served as the prototypes for a diverse collection of interaction domains that recognize not only proteins but also lipids, nucleic acids, and small molecules. It has also been found that particular SH2 and SH3 domains themselves might also bind to and rely on lipids to modulate complex assembly. Some lipid-binding properties of SH2 and SH3 domains are reviewed here.
Keywords: SH2 domain; SH3 domain; Src; lipid binding; proline-rich sequences; protein tyrosine kinase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Margolis B., Li N., Koch A., Mohammadi M., Hurwitz D.R., Zilberstein A., Ullrich A., Pawson T., Schlessinger J. The tyrosine phosphorylated carboxyterminus of the EGF receptor is a binding site for GAP and PLC-γ. EMBO J. 1990;9:4375–4380. doi: 10.1002/j.1460-2075.1990.tb07887.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

