Iron in Translation: From the Beginning to the End
- PMID: 34068342
- PMCID: PMC8153317
- DOI: 10.3390/microorganisms9051058
Iron in Translation: From the Beginning to the End
Abstract
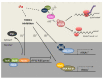
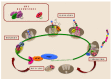
Iron is an essential element for all eukaryotes, since it acts as a cofactor for many enzymes involved in basic cellular functions, including translation. While the mammalian iron-regulatory protein/iron-responsive element (IRP/IRE) system arose as one of the first examples of translational regulation in higher eukaryotes, little is known about the contribution of iron itself to the different stages of eukaryotic translation. In the yeast Saccharomyces cerevisiae, iron deficiency provokes a global impairment of translation at the initiation step, which is mediated by the Gcn2-eIF2α pathway, while the post-transcriptional regulator Cth2 specifically represses the translation of a subgroup of iron-related transcripts. In addition, several steps of the translation process depend on iron-containing enzymes, including particular modifications of translation elongation factors and transfer RNAs (tRNAs), and translation termination by the ATP-binding cassette family member Rli1 (ABCE1 in humans) and the prolyl hydroxylase Tpa1. The influence of these modifications and their correlation with codon bias in the dynamic control of protein biosynthesis, mainly in response to stress, is emerging as an interesting focus of research. Taking S. cerevisiae as a model, we hereby discuss the relevance of iron in the control of global and specific translation steps.
Keywords: Saccharomyces cerevisiae; iron deficiency; tRNA modification; translation; yeast.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

