Differential Methylation Patterns in Apomictic vs. Sexual Genotypes of the Diplosporous Grass Eragrostis curvula
- PMID: 34068493
- PMCID: PMC8150776
- DOI: 10.3390/plants10050946
Differential Methylation Patterns in Apomictic vs. Sexual Genotypes of the Diplosporous Grass Eragrostis curvula
Abstract
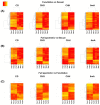
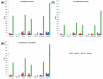
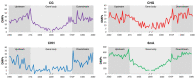
DNA methylation is an epigenetic mechanism by which a methyl group is added to a cytosine or an adenine. When located in a gene/regulatory sequence it may repress or de-repress genes, depending on the context and species. Eragrostis curvula is an apomictic grass in which facultative genotypes increases the frequency of sexual pistils triggered by epigenetic mechanisms. The aim of the present study was to look for correlations between the reproductive mode and specific methylated genes or genomic regions. To do so, plants with contrasting reproductive modes were investigated through MCSeEd (Methylation Context Sensitive Enzyme ddRad) showing higher levels of DNA methylation in apomictic genotypes. Moreover, an increased proportion of differentially methylated positions over the regulatory regions were observed, suggesting its possible role in regulation of gene expression. Interestingly, the methylation pathway was also found to be self-regulated since two of the main genes (ROS1 and ROS4), involved in de-methylation, were found differentially methylated between genotypes with different reproductive behavior. Moreover, this work allowed us to detect several genes regulated by methylation that were previously found as differentially expressed in the comparisons between apomictic and sexual genotypes, linking DNA methylation to differences in reproductive mode.
Keywords: DNA methylation; apomixis; diplospory; epigenetics; eragrostis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

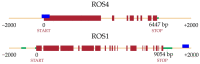

Similar articles
-
Differentially methylated genes involved in reproduction and ploidy levels in recent diploidized and tetraploidized Eragrostis curvula genotypes.Plant Reprod. 2024 Jun;37(2):133-145. doi: 10.1007/s00497-023-00490-7. Epub 2023 Dec 6. Plant Reprod. 2024. PMID: 38055074 Free PMC article.
-
New insights into Eragrostis curvula's sexual and apomictic reproductive development.Front Plant Sci. 2025 May 1;16:1530855. doi: 10.3389/fpls.2025.1530855. eCollection 2025. Front Plant Sci. 2025. PMID: 40376162 Free PMC article.
-
Genes Modulating the Increase in Sexuality in the Facultative Diplosporous Grass Eragrostis curvula under Water Stress Conditions.Genes (Basel). 2020 Aug 21;11(9):969. doi: 10.3390/genes11090969. Genes (Basel). 2020. PMID: 32825586 Free PMC article.
-
Eragrostis curvula, a Model Species for Diplosporous Apomixis.Plants (Basel). 2021 Aug 31;10(9):1818. doi: 10.3390/plants10091818. Plants (Basel). 2021. PMID: 34579351 Free PMC article. Review.
-
Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms.Curr Opin Plant Biol. 2012 Feb;15(1):57-62. doi: 10.1016/j.pbi.2011.10.002. Epub 2011 Oct 28. Curr Opin Plant Biol. 2012. PMID: 22037465 Review.
Cited by
-
A sexual/apomictic consensus linkage map of Eragrostis curvula at tetraploid level.BMC Plant Biol. 2025 May 17;25(1):658. doi: 10.1186/s12870-025-06676-7. BMC Plant Biol. 2025. PMID: 40382602 Free PMC article.
-
Genetic Transformation of Apomictic Grasses: Progress and Constraints.Front Plant Sci. 2021 Nov 5;12:768393. doi: 10.3389/fpls.2021.768393. eCollection 2021. Front Plant Sci. 2021. PMID: 34804102 Free PMC article. Review.
-
From tetraploid to diploid, a pangenomic approach to identify genes lost during synthetic diploidization of Eragrostis curvula.Front Plant Sci. 2023 Mar 13;14:1133986. doi: 10.3389/fpls.2023.1133986. eCollection 2023. Front Plant Sci. 2023. PMID: 36993842 Free PMC article.
-
Differentially methylated genes involved in reproduction and ploidy levels in recent diploidized and tetraploidized Eragrostis curvula genotypes.Plant Reprod. 2024 Jun;37(2):133-145. doi: 10.1007/s00497-023-00490-7. Epub 2023 Dec 6. Plant Reprod. 2024. PMID: 38055074 Free PMC article.
-
New insights into Eragrostis curvula's sexual and apomictic reproductive development.Front Plant Sci. 2025 May 1;16:1530855. doi: 10.3389/fpls.2025.1530855. eCollection 2025. Front Plant Sci. 2025. PMID: 40376162 Free PMC article.
References
-
- Russo V.E., Martienssen R.A., Riggs A.D. Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press; Long Island, NY, USA: 1996.
Grants and funding
LinkOut - more resources
Full Text Sources

