Viruses with U-DNA: New Avenues for Biotechnology
- PMID: 34068736
- PMCID: PMC8150378
- DOI: 10.3390/v13050875
Viruses with U-DNA: New Avenues for Biotechnology
Abstract
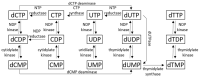
Deoxyuridine in DNA has recently been in the focus of research due to its intriguing roles in several physiological and pathophysiological situations. Although not an orthodox DNA base, uracil may appear in DNA via either cytosine deamination or thymine-replacing incorporations. Since these alterations may induce mutation or may perturb DNA-protein interactions, free living organisms from bacteria to human contain several pathways to counteract uracilation. These efficient and highly specific repair routes uracil-directed excision repair initiated by representative of uracil-DNA glycosylase families. Interestingly, some bacteriophages exist with thymine-lacking uracil-DNA genome. A detailed understanding of the strategy by which such phages can replicate in bacteria where an efficient repair pathway functions for uracil-excision from DNA is expected to reveal novel inhibitors that can also be used for biotechnological applications. Here, we also review the several potential biotechnological applications already implemented based on inhibitors of uracil-excision repair, such as Crispr-base-editing and detection of nascent uracil distribution pattern in complex genomes.
Keywords: biotechnology; phages; uracil-DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Róna G., Marfori M., Borsos M., Scheer I., Takács E., Tóth J., Babos F., Magyar A., Erdei A., Bozóky Z., et al. Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: Structural and mechanistic insights. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:2495–2505. doi: 10.1107/S0907444913023354. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

