The Mechanism of Asparagine Endopeptidase in the Progression of Malignant Tumors: A Review
- PMID: 34068767
- PMCID: PMC8151911
- DOI: 10.3390/cells10051153
The Mechanism of Asparagine Endopeptidase in the Progression of Malignant Tumors: A Review
Abstract
Asparagine endopeptidase (AEP), also called legumain, is currently the only known cysteine protease that specifically cleaves peptide bonds in asparaginyl residue in the mammalian genome. Since 2003, AEP has been reported to be widely expressed in a variety of carcinomas and is considered a potential therapeutic target. In the following years, researchers intensively investigated the substrates of AEP and the mechanism of AEP in partial tumors. With the identification of substrate proteins such as P53, integrin αvβ3, MMP-2, and MMP-9, the biochemical mechanism of AEP in carcinomas is also more precise. This review will clarify the probable mechanisms of AEP in the progression of breast carcinoma, glioblastoma, gastric carcinoma, and epithelial ovarian carcinoma. This review will also discuss the feasibility of targeted therapy with AEP inhibitor (AEPI) in these carcinomas.
Keywords: asparagine endopeptidase; breast cancer; epithelial ovarian cancer; gastric cancer; glioblastoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
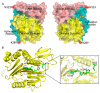

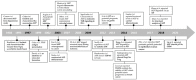
Similar articles
-
The expression of asparaginyl endopeptidase promotes growth potential in epithelial ovarian cancer.Cancer Biol Ther. 2017 Apr 3;18(4):222-228. doi: 10.1080/15384047.2017.1294290. Epub 2017 Mar 3. Cancer Biol Ther. 2017. PMID: 28278071 Free PMC article.
-
Role of Asparagine Endopeptidase in Mediating Wild-Type p53 Inactivation of Glioblastoma.J Natl Cancer Inst. 2020 Apr 1;112(4):343-355. doi: 10.1093/jnci/djz155. J Natl Cancer Inst. 2020. PMID: 31400201 Free PMC article.
-
Asparaginyl endopeptidase promotes the invasion and metastasis of gastric cancer through modulating epithelial-to-mesenchymal transition and analysis of their phosphorylation signaling pathways.Oncotarget. 2016 Jun 7;7(23):34356-70. doi: 10.18632/oncotarget.8879. Oncotarget. 2016. PMID: 27102302 Free PMC article.
-
Asparagine endopeptidase is an innovative therapeutic target for neurodegenerative diseases.Expert Opin Ther Targets. 2016 Oct;20(10):1237-45. doi: 10.1080/14728222.2016.1182990. Epub 2016 May 13. Expert Opin Ther Targets. 2016. PMID: 27115710 Free PMC article. Review.
-
Development of asparagine endopeptidase inhibitors for treating neurodegenerative diseases.Trends Mol Med. 2025 Apr;31(4):359-372. doi: 10.1016/j.molmed.2025.01.009. Epub 2025 Feb 25. Trends Mol Med. 2025. PMID: 40000317 Review.
Cited by
-
The Mammalian Cysteine Protease Legumain in Health and Disease.Int J Mol Sci. 2022 Dec 15;23(24):15983. doi: 10.3390/ijms232415983. Int J Mol Sci. 2022. PMID: 36555634 Free PMC article. Review.
-
Proteomic Identification of a Gastric Tumor ECM Signature Associated With Cancer Progression.Front Mol Biosci. 2022 Mar 1;9:818552. doi: 10.3389/fmolb.2022.818552. eCollection 2022. Front Mol Biosci. 2022. PMID: 35340765 Free PMC article.
-
The Proteolytic Landscape of Ovarian Cancer: Applications in Nanomedicine.Int J Mol Sci. 2022 Sep 1;23(17):9981. doi: 10.3390/ijms23179981. Int J Mol Sci. 2022. PMID: 36077371 Free PMC article. Review.
-
Radiotherapy resistance driven by Asparagine endopeptidase through ATR pathway modulation in breast cancer.J Exp Clin Cancer Res. 2025 Feb 27;44(1):74. doi: 10.1186/s13046-025-03334-6. J Exp Clin Cancer Res. 2025. PMID: 40012043 Free PMC article.
-
Lysosomal peptidases-intriguing roles in cancer progression and neurodegeneration.FEBS Open Bio. 2022 Apr;12(4):708-738. doi: 10.1002/2211-5463.13372. Epub 2022 Feb 3. FEBS Open Bio. 2022. PMID: 35067006 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

