Microbial Composition of SCOBY Starter Cultures Used by Commercial Kombucha Brewers in North America
- PMID: 34068887
- PMCID: PMC8156240
- DOI: 10.3390/microorganisms9051060
Microbial Composition of SCOBY Starter Cultures Used by Commercial Kombucha Brewers in North America
Abstract
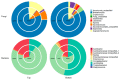
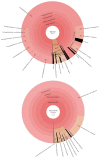
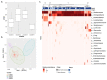
Kombucha fermentation is initiated by transferring a solid-phase cellulosic pellicle into sweetened tea and allowing the microbes that it contains to initiate the fermentation. This pellicle, commonly referred to as a symbiotic culture of bacteria and yeast (SCOBY), floats to the surface of the fermenting tea and represents an interphase environment, where embedded microbes gain access to oxygen as well as nutrients in the tea. To date, various yeast and bacteria have been reported to exist within the SCOBY, with little consensus as to which species are essential and which are incidental to Kombucha production. In this study, we used high-throughput sequencing approaches to evaluate spatial homogeneity within a single commercial SCOBY and taxonomic diversity across a large number (n = 103) of SCOBY used by Kombucha brewers, predominantly in North America. Our results show that the most prevalent and abundant SCOBY taxa were the yeast genus Brettanomyces and the bacterial genus Komagataeibacter, through careful sampling of upper and lower SCOBY layers. This sampling procedure is critical to avoid over-representation of lactic acid bacteria. K-means clustering was used on metabarcoding data of all 103 SCOBY, delineating four SCOBY archetypes based upon differences in their microbial community structures. Fungal genera Zygosaccharomyces, Lachancea and Starmerella were identified as the major compensatory taxa for SCOBY with lower relative abundance of Brettanomyces. Interestingly, while Lactobacillacae was the major compensatory taxa where Komagataeibacter abundance was lower, phylogenic heat-tree analysis infers a possible antagonistic relationship between Starmerella and the acetic acid bacterium. Our results provide the basis for further investigation of how SCOBY archetype affects Kombucha fermentation, and fundamental studies of microbial community assembly in an interphase environment.
Keywords: Brettanomyces; Illumina sequencing; Kombucha fermentation; acetic acid bacteria; microbiome; mycobiome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Malbaša R.V., Lončar E.S., Vitas J.S., Čanadanović-Brunet J.M. Influence of starter cultures on the antioxidant activity of kombucha beverage. Food Chem. 2011;127:1727–1731. doi: 10.1016/j.foodchem.2011.02.048. - DOI
-
- Yamada Y., Yukphan P., Vu H.T.L., Muramatsu Y., Ochaikul D., Nakagawa Y. Subdivision of the Genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: The Proposal of Komagataeibacter Gen. Nov., for Strains Accommodated to the Gluconacetobacter Xylinus Group in the α-Proteobacteria. Ann. Microbiol. 2012;62:849–859. doi: 10.1007/s13213-011-0288-4. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

