Development of New PCR Assay with SYBR Green I for Detection of Mycoplasma, Acholeplasma, and Ureaplasma sp. in Cell Cultures
- PMID: 34068904
- PMCID: PMC8156504
- DOI: 10.3390/diagnostics11050876
Development of New PCR Assay with SYBR Green I for Detection of Mycoplasma, Acholeplasma, and Ureaplasma sp. in Cell Cultures
Abstract
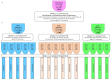
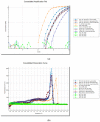
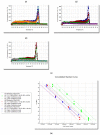
Mycoplasma, Acholeplasma, and Ureaplasma sp. are atypical bacteria responsible for in vitro cell culture contaminations that can warp the results. These bacteria also cause human and animal infections and may lead to chronic diseases. In developed polymerase chain reaction (PCR) in this study a quantitative PCR with SYBR Green I fluorochrome was applied to facilitate the Mycoplasma, Acholeplasma, and Ureaplasma sp. DNA detection and identification. Screening Test-1 v.1 (triplex qPCR) allowed for the detection of 11 species. Test-1 v.2 (three single qPCRs) pre-identified three subgroups, allowing for the reduction of using single qPCRs in Test-2 for species identification. The range of both tests was consistent with pharmacopeial requirements for microbial quality control of mammal cells and included detection of M. arginini, M. orale, M. hyorhinis, M. fermentans, M. genitalium, M. hominis, M. pneumoniae, M. salivarium, M. pirum, A. laidlawii, and U. urealyticum. Limit of detection values varied between 125-300 and 50-100 number of copies per milliliter in Test-1 and Test-2, respectively. Test-1 and Test-2 showed fully concordant results, allowed for time-saving detection and/or identification of selected species from Mycoplasma, Acholeplasma, and Ureaplasma in tested cell cultures.
Keywords: Acholeplasma and Ureaplasma sp. detection; Mycoplasma; cell cultures; qPCR validation; quality control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Simultaneous detection and identification of common cell culture contaminant and pathogenic mollicutes strains by reverse line blot hybridization.Appl Environ Microbiol. 2004 Mar;70(3):1483-6. doi: 10.1128/AEM.70.3.1483-1486.2004. Appl Environ Microbiol. 2004. PMID: 15006769 Free PMC article.
-
A polymerase chain reaction based method for detecting Mycoplasma/Acholeplasma contaminants in cell culture.J Microbiol Methods. 2000 Jan;39(2):121-6. doi: 10.1016/s0167-7012(99)00107-4. J Microbiol Methods. 2000. PMID: 10576701
-
Rapid detection of mycoplasma contamination in cell cultures using SYBR Green-based real-time polymerase chain reaction.In Vitro Cell Dev Biol Anim. 2006 Mar-Apr;42(3-4):63-9. doi: 10.1290/0505035.1. In Vitro Cell Dev Biol Anim. 2006. PMID: 16759150
-
Genital mycoplasma infections.Wien Klin Wochenschr. 1997 Aug 8;109(14-15):578-83. Wien Klin Wochenschr. 1997. PMID: 9286063 Review.
-
DNA probes and PCR in diagnosis of mycoplasma infections.Mol Cell Probes. 1994 Dec;8(6):497-511. doi: 10.1006/mcpr.1994.1071. Mol Cell Probes. 1994. PMID: 7700272 Review.
Cited by
-
Comparative genomic analysis of Mycoplasma related to cell culture for infB gene-based loop-mediated isothermal amplification.World J Microbiol Biotechnol. 2023 Oct 25;39(12):355. doi: 10.1007/s11274-023-03794-y. World J Microbiol Biotechnol. 2023. PMID: 37878143
-
The associations between low abundance of Mycoplasma hominis and female fecundability: a pregnancy-planning cohort study.BMC Microbiol. 2022 May 5;22(1):121. doi: 10.1186/s12866-022-02545-7. BMC Microbiol. 2022. PMID: 35513786 Free PMC article.
References
-
- Volokhov D.V., Simonyan V., Davidson M.K., Chizhikov V.E. RNA polymerase beta subunit (rpoB) gene and the 16S-23S rRNA intergenic transcribed spacer region (ITS) as complementary molecular markers in addition to the 16S rRNA gene for phylogenetic analysis and identification of the species of the family Mycoplasmataceae. Mol. Phylogenet. Evol. 2012;62:515–528. doi: 10.1016/j.ympev.2011.11.002. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

