Carotenoid Nostoxanthin Production by Sphingomonas sp. SG73 Isolated from Deep Sea Sediment
- PMID: 34068940
- PMCID: PMC8156329
- DOI: 10.3390/md19050274
Carotenoid Nostoxanthin Production by Sphingomonas sp. SG73 Isolated from Deep Sea Sediment
Abstract
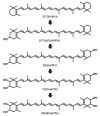
Carotenoids are used commercially for dietary supplements, cosmetics, and pharmaceuticals because of their antioxidant activity. In this study, colored microorganisms were isolated from deep sea sediment that had been collected from Suruga Bay, Shizuoka, Japan. One strain was found to be a pure yellow carotenoid producer, and the strain was identified as Sphingomonas sp. (Proteobacteria) by 16S rRNA gene sequence analysis; members of this genus are commonly isolated from air, the human body, and marine environments. The carotenoid was identified as nostoxanthin ((2,3,2',3')-β,β-carotene-2,3,2',3'-tetrol) by mass spectrometry (MS), MS/MS, and ultraviolet-visible absorption spectroscopy (UV-Vis). Nostoxanthin is a poly-hydroxy yellow carotenoid isolated from some photosynthetic bacteria, including some species of Cyanobacteria. The strain Sphingomonas sp. SG73 produced highly pure nostoxanthin of approximately 97% (area%) of the total carotenoid production, and the strain was halophilic and tolerant to 1.5-fold higher salt concentration as compared with seawater. When grown in 1.8% artificial sea salt, nostoxanthin production increased by 2.5-fold as compared with production without artificial sea salt. These results indicate that Sphingomonas sp. SG73 is an efficient producer of nostoxanthin, and the strain is ideal for carotenoid production using marine water because of its compatibility with sea salt.
Keywords: Sphingomonas; carotenoid; deep sea microorganism; nostoxanthin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Nostoxanthin Biosynthesis by Sphingomonas Species (COS14-R2): Isolation, Identification, and Optimization of Culture Conditions.Curr Microbiol. 2024 Nov 11;81(12):453. doi: 10.1007/s00284-024-03956-7. Curr Microbiol. 2024. PMID: 39527245 Free PMC article.
-
Sphingomonas jaspsi sp. nov., a novel carotenoid-producing bacterium isolated from Misasa, Tottori, Japan.Int J Syst Evol Microbiol. 2007 Jul;57(Pt 7):1435-1441. doi: 10.1099/ijs.0.64828-0. Int J Syst Evol Microbiol. 2007. PMID: 17625171
-
Sphingomonas profundi sp. nov., isolated from deep-sea sediment of the Mariana Trench.Int J Syst Evol Microbiol. 2020 Jun;70(6):3809-3815. doi: 10.1099/ijsem.0.004235. Epub 2020 Jun 4. Int J Syst Evol Microbiol. 2020. PMID: 32496177
-
Isolation of a beta-carotene over-producing soil bacterium, Sphingomonas sp.Biotechnol Lett. 2004 Feb;26(3):257-62. doi: 10.1023/b:bile.0000013716.20116.dc. Biotechnol Lett. 2004. PMID: 15049373
-
Cloning and characterization of genes involved in nostoxanthin biosynthesis of Sphingomonas elodea ATCC 31461.PLoS One. 2012;7(4):e35099. doi: 10.1371/journal.pone.0035099. Epub 2012 Apr 11. PLoS One. 2012. PMID: 22509387 Free PMC article.
Cited by
-
Characterization and Bioactive Potential of Carotenoid Lutein from Gordonia rubripertncta GH-1 Isolated from Traditional Pixian Douban.Foods. 2022 Nov 15;11(22):3649. doi: 10.3390/foods11223649. Foods. 2022. PMID: 36429243 Free PMC article.
-
A nostoxanthin-producing bacterium, Sphingomonas nostoxanthinifaciens sp. nov., alleviates the salt stress of Arabidopsis seedlings by scavenging of reactive oxygen species.Front Microbiol. 2023 Feb 10;14:1101150. doi: 10.3389/fmicb.2023.1101150. eCollection 2023. Front Microbiol. 2023. PMID: 36846770 Free PMC article.
-
Metabolomic Analysis of Carotenoids Biosynthesis by Sphingopyxis sp. USTB-05.Molecules. 2024 Sep 6;29(17):4235. doi: 10.3390/molecules29174235. Molecules. 2024. PMID: 39275082 Free PMC article.
-
Nostoxanthin Biosynthesis by Sphingomonas Species (COS14-R2): Isolation, Identification, and Optimization of Culture Conditions.Curr Microbiol. 2024 Nov 11;81(12):453. doi: 10.1007/s00284-024-03956-7. Curr Microbiol. 2024. PMID: 39527245 Free PMC article.
-
Limnospira (Cyanobacteria) chemical fingerprint reveals local molecular adaptation.Microbiol Spectr. 2025 Feb 4;13(2):e0190124. doi: 10.1128/spectrum.01901-24. Epub 2025 Jan 8. Microbiol Spectr. 2025. PMID: 39772964 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

