High Flexibility of RNaseH2 Catalytic Activity with Respect to Non-Canonical DNA Structures
- PMID: 34068992
- PMCID: PMC8155979
- DOI: 10.3390/ijms22105201
High Flexibility of RNaseH2 Catalytic Activity with Respect to Non-Canonical DNA Structures
Abstract
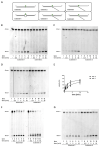
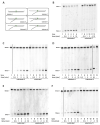
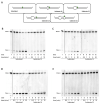
Ribonucleotides misincorporated in the human genome are the most abundant DNA lesions. The 2'-hydroxyl group makes them prone to spontaneous hydrolysis, potentially resulting in strand breaks. Moreover, their presence may decrease the rate of DNA replication causing replicative fork stalling and collapse. Ribonucleotide removal is initiated by Ribonuclease H2 (RNase H2), the key player in Ribonucleotide Excision Repair (RER). Its absence leads to embryonic lethality in mice, while mutations decreasing its activity cause Aicardi-Goutières syndrome. DNA geometry can be altered by DNA lesions or by peculiar sequences forming secondary structures, like G-quadruplex (G4) and trinucleotide repeats (TNR) hairpins, which significantly differ from canonical B-form. Ribonucleotides pairing to lesioned nucleotides, or incorporated within non-B DNA structures could avoid RNase H2 recognition, potentially contributing to genome instability. In this work, we investigate the ability of RNase H2 to process misincorporated ribonucleotides in a panel of DNA substrates showing different geometrical features. RNase H2 proved to be a flexible enzyme, recognizing as a substrate the majority of the constructs we generated. However, some geometrical features and non-canonical DNA structures severely impaired its activity, suggesting a relevant role of misincorporated ribonucleotides in the physiological instability of specific DNA sequences.
Keywords: RER; RNaseH2; misincorporated ribonucleotides; non-B DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
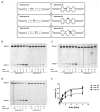
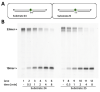
References
-
- Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundström E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. - DOI - PMC - PubMed
-
- Ghodgaonkar M.M., Lazzaro F., Olivera-Pimentel M., Artola-Borán M., Cejka P., Reijns M.A., Jackson A.P., Plevani P., Muzi-Falconi M., Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

