New Approach for Untangling the Role of Uncommon Calcium-Binding Proteins in the Central Nervous System
- PMID: 34069107
- PMCID: PMC8156796
- DOI: 10.3390/brainsci11050634
New Approach for Untangling the Role of Uncommon Calcium-Binding Proteins in the Central Nervous System
Abstract
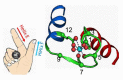
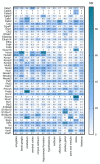
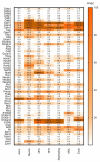
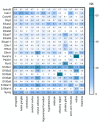
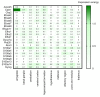
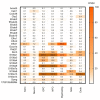
Although Ca2+ ion plays an essential role in cellular physiology, calcium-binding proteins (CaBPs) were long used for mainly as immunohistochemical markers of specific cell types in different regions of the central nervous system. They are a heterogeneous and wide-ranging group of proteins. Their function was studied intensively in the last two decades and a tremendous amount of information was gathered about them. Girard et al. compiled a comprehensive list of the gene-expression profiles of the entire EF-hand gene superfamily in the murine brain. We selected from this database those CaBPs which are related to information processing and/or neuronal signalling, have a Ca2+-buffer activity, Ca2+-sensor activity, modulator of Ca2+-channel activity, or a yet unknown function. In this way we created a gene function-based selection of the CaBPs. We cross-referenced these findings with publicly available, high-quality RNA-sequencing and in situ hybridization databases (Human Protein Atlas (HPA), Brain RNA-seq database and Allen Brain Atlas integrated into the HPA) and created gene expression heat maps of the regional and cell type-specific expression levels of the selected CaBPs. This represents a useful tool to predict and investigate different expression patterns and functions of the less-known CaBPs of the central nervous system.
Keywords: calcium-binding proteins; central nervous system; human protein atlas; in situ hybridisation database; transcriptome database.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The EF-hand Ca(2+)-binding protein super-family: a genome-wide analysis of gene expression patterns in the adult mouse brain.Neuroscience. 2015 May 21;294:116-55. doi: 10.1016/j.neuroscience.2015.02.018. Epub 2015 Mar 12. Neuroscience. 2015. PMID: 25770968
-
Structural and functional diversity of Entamoeba histolytica calcium-binding proteins.Biophys Rev. 2020 Oct 15;12(6):1331-41. doi: 10.1007/s12551-020-00766-6. Online ahead of print. Biophys Rev. 2020. PMID: 33063237 Free PMC article. Review.
-
Evolution and functional diversity of the Calcium Binding Proteins (CaBPs).Front Mol Neurosci. 2012 Feb 21;5:9. doi: 10.3389/fnmol.2012.00009. eCollection 2012 Jan 23. Front Mol Neurosci. 2012. PMID: 22375103 Free PMC article.
-
Differential transcriptional expression of Ca2+ BP superfamilies in murine gastrointestinal smooth muscles.Am J Physiol Gastrointest Liver Physiol. 2002 Dec;283(6):G1290-7. doi: 10.1152/ajpgi.00101.2002. Epub 2002 May 29. Am J Physiol Gastrointest Liver Physiol. 2002. PMID: 12388203
-
Calcium binding proteins and calcium signaling in prokaryotes.Cell Calcium. 2015 Mar;57(3):151-65. doi: 10.1016/j.ceca.2014.12.006. Epub 2014 Dec 17. Cell Calcium. 2015. PMID: 25555683 Review.
Cited by
-
Distribution of NECAB1-Positive Neurons in Normal and Epileptic Brain-Expression Changes in Temporal Lobe Epilepsy and Modulation by Levetiracetam and Brivaracetam.Int J Mol Sci. 2025 May 20;26(10):4906. doi: 10.3390/ijms26104906. Int J Mol Sci. 2025. PMID: 40430045 Free PMC article.
-
Simultaneous RNA Sequencing and DNA Methylation Profiling Reveals Neural Mechanisms That Regulate Sensitive Period Behavioral Learning.Genes Brain Behav. 2025 Aug;24(4):e70031. doi: 10.1111/gbb.70031. Genes Brain Behav. 2025. PMID: 40776851 Free PMC article.
-
Patient-Specific iPSCs-Based Models of Neurodegenerative Diseases: Focus on Aberrant Calcium Signaling.Int J Mol Sci. 2022 Jan 6;23(2):624. doi: 10.3390/ijms23020624. Int J Mol Sci. 2022. PMID: 35054808 Free PMC article. Review.
-
Rabies virus infection is associated with variations in calbindin D-28K and calretinin mRNA expression levels in mouse brain tissue.Arch Virol. 2023 Apr 18;168(5):143. doi: 10.1007/s00705-023-05753-2. Arch Virol. 2023. PMID: 37069450 Free PMC article.
-
Characterization and Extraction Influence Protein Profiling of Edible Bird's Nest.Foods. 2021 Sep 23;10(10):2248. doi: 10.3390/foods10102248. Foods. 2021. PMID: 34681297 Free PMC article.
References
-
- Kandel E.R., Schwartz J.H., Jessell T.M., Siegelbaum S.A., Hudspeth A.J. Principles of Neural Science. 5th ed. McGraw-Hill; New York, NY, USA: 2013. Health Professions Division.
-
- Lodish H. Molecular Cell Biology. 8th ed. W.H. Freeman; New York, NY, USA: 2016.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

