Transcriptional Profiling of Porcine Blastocysts Produced In Vitro in a Chemically Defined Culture Medium
- PMID: 34069238
- PMCID: PMC8156047
- DOI: 10.3390/ani11051414
Transcriptional Profiling of Porcine Blastocysts Produced In Vitro in a Chemically Defined Culture Medium
Abstract
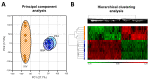
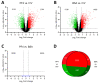
The development of chemically defined media is a growing trend in in vitro embryo production (IVP). Recently, traditional undefined culture medium with bovine serum albumin (BSA) has been successfully replaced by a chemically defined medium using substances with embryotrophic properties such as platelet factor 4 (PF4). Although the use of this medium sustains IVP, the impact of defined media on the embryonic transcriptome has not been fully elucidated. This study analyzed the transcriptome of porcine IVP blastocysts, cultured in defined (PF4 group) and undefined media (BSA group) by microarrays. In vivo-derived blastocysts (IVV group) were used as a standard of maximum embryo quality. The results showed no differentially expressed genes (DEG) between the PF4 and BSA groups. However, a total of 2780 and 2577 DEGs were detected when comparing the PF4 or the BSA group with the IVV group, respectively. Most of these genes were common in both in vitro groups (2132) and present in some enriched pathways, such as cell cycle, lysosome and/or metabolic pathways. These results show that IVP conditions strongly affect embryo transcriptome and that the defined culture medium with PF4 is a guaranteed replacement for traditional culture with BSA.
Keywords: BSA; PF4; defined media; embryo; in vitro; microarray; porcine; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






Similar articles
-
The cytokine platelet factor 4 successfully replaces bovine serum albumin for the in vitro culture of porcine embryos.Theriogenology. 2020 May;148:201-207. doi: 10.1016/j.theriogenology.2019.11.009. Epub 2019 Nov 10. Theriogenology. 2020. PMID: 31748174
-
Improved in vitro bovine embryo development and increased efficiency in producing viable calves using defined media.Theriogenology. 2007 Jan 15;67(2):293-302. doi: 10.1016/j.theriogenology.2006.07.011. Epub 2006 Sep 15. Theriogenology. 2007. PMID: 16979228
-
High bovine blastocyst development in a static in vitro production system using SOFaa medium supplemented with sodium citrate and myo-inositol with or without serum-proteins.Theriogenology. 1999 Sep;52(4):683-700. doi: 10.1016/S0093-691X(99)00162-4. Theriogenology. 1999. PMID: 10734366
-
Bovine embryo culture in the presence or absence of serum: implications for blastocyst development, cryotolerance, and messenger RNA expression.Biol Reprod. 2003 Jan;68(1):236-43. doi: 10.1095/biolreprod.102.007799. Biol Reprod. 2003. PMID: 12493719
-
Development and application of a chemically defined medium for the in vitro production of porcine embryos.J Reprod Dev. 2011 Feb;57(1):9-16. doi: 10.1262/jrd.10-196e. J Reprod Dev. 2011. PMID: 21422733 Review.
Cited by
-
Oviductal Extracellular Vesicles Enhance Porcine In Vitro Embryo Development by Modulating the Embryonic Transcriptome.Biomolecules. 2022 Sep 15;12(9):1300. doi: 10.3390/biom12091300. Biomolecules. 2022. PMID: 36139139 Free PMC article.
References
-
- Whitworth K.M., Agca C., Kim J.-G., Patel R.V., Springer G.K., Bivens N.J., Forrester L.J., Mathialagan N., Green J.A., Prather R.S. Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol. Reprod. 2005;72:1437–1451. doi: 10.1095/biolreprod.104.037952. - DOI - PubMed
-
- Miles J.R., Blomberg L.A., Krisher R.L., Everts R.E., Sonstegard T.S., Van Tassell C.P., Zuelke K.A. Comparative transcriptome analysis of in vivo- and in vitro-produced porcine blastocysts by small amplified RNA-serial analysis of gene expression (SAR-SAGE) Mol. Reprod. Dev. 2008;75:976–988. doi: 10.1002/mrd.20844. - DOI - PubMed
-
- Bauer B.K., Isom S.C., Spate L.D., Whitworth K.M., Spollen W.G., Blake S.M., Springer G.K., Murphy C.N., Prather R.S. Transcriptional profiling by deep sequencing identifies differences in mRNA transcript abundance in in vivo-derived versus in vitro-cultured porcine blastocyst stage embryos. Biol. Reprod. 2010;83:791–798. doi: 10.1095/biolreprod.110.085936. - DOI - PubMed
-
- Canovas S., Ivanova E., Romar R., García-Martínez S., Soriano-Úbeda C., García-Vázquez F.A., Saadeh H., Andrews S., Kelsey G., Coy P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. Elife. 2017;6 doi: 10.7554/eLife.23670. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

