M448R and MGF505-7R: Two African Swine Fever Virus Antigens Commonly Recognized by ASFV-Specific T-Cells and with Protective Potential
- PMID: 34069239
- PMCID: PMC8156282
- DOI: 10.3390/vaccines9050508
M448R and MGF505-7R: Two African Swine Fever Virus Antigens Commonly Recognized by ASFV-Specific T-Cells and with Protective Potential
Abstract
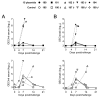
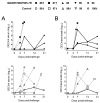
African swine fever (ASF) is today's number one threat for the global swine industry. Neither commercial vaccine nor treatment is available against ASF and, thus far, only live attenuated viruses (LAV) have provided robust protection against lethal ASF virus (ASFV) challenge infections. Identification of ASFV proteins inducing protective immune responses is one of the major challenges to develop safer and efficient subunit vaccines. Immunopeptidomic studies recently performed in our laboratory allowed identifying ASFV antigens recognized by ASFV-specific CD8+ T-cells. Here, we used data from the SLAI-peptide repertoire presented by a single set of ASFV-infected porcine alveolar macrophages to generate a complex DNA vaccine composed by 15 plasmids encoding the individual peptide-bearing ORFs. DNA vaccine priming improved the protection afforded by a suboptimal dose of the BA71ΔCD2 LAV given as booster vaccination, against Georgia2007/1 lethal challenge. Interestingly, M448R was the only protein promiscuously recognized by the induced ASFV-specific T-cells. Furthermore, priming pigs with DNA plasmids encoding M488R and MGF505-7R, a CD8+ T-cell antigen previously described, confirmed these two proteins as T-cell antigens with protective potential. These studies might be useful to pave the road for designing safe and more efficient vaccine formulations in the future.
Keywords: African swine fever; DNA immunization; T-cells; antigen discovery; immunopeptidomics; live attenuated virus; protection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Beltran-Alcrudo D., Lubroth J., Depner K., La Rocque S. African swine fever in the Caucasus. FAO Empres. Watch. 2008;1:1–8.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

